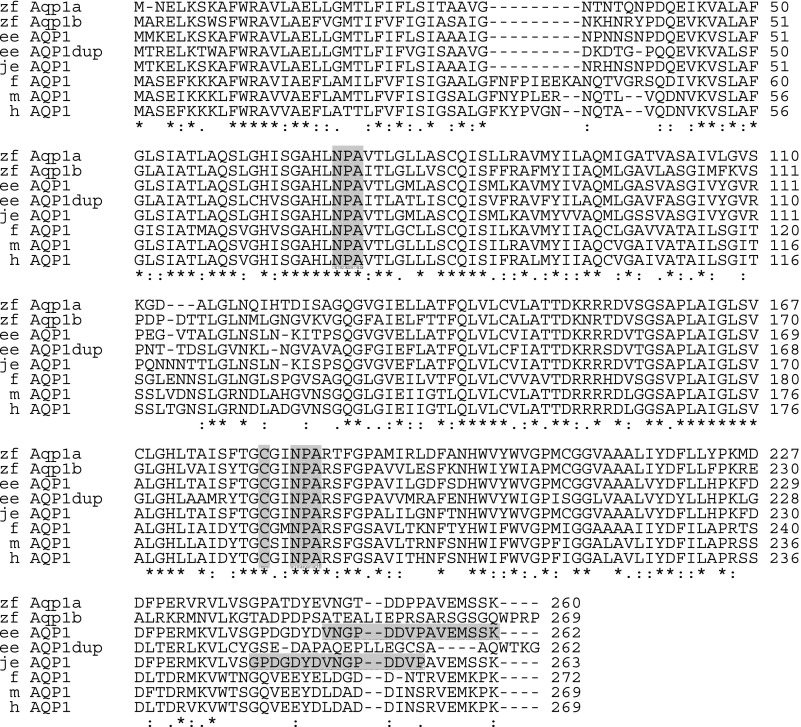
Fig. 2.
Alignment of aquaporin 1 (AQP1) sequences from zebrafish and other selected species. Sequence alignment was performed using the online program ClustalW of European Bioinformatics Institute (EBI; http://www.ebi.ac.uk/clustalw/) with default gap parameters (GapOpen = 10.0, EndGap = −1, GapExtension = 0.2, and GapDistance = 4). GenBank accession numbers of the sequences analyzed are given in parentheses: zebrafish (zf, Danio rerio) aqp1a (DQ887675) and aqp1b (EU327345); European eel (ee, Anguilla anguilla) AQP1 (AJ564420) and AQP1dup (AJ564421); Japanese eel (je, Anguilla japonica) AQP1 (AB094501); frog (f, Rana esculenta) AQP1 (L24754); mouse (m, Mus musculus) AQP1 (BC007125); and human (h, homo sapiens) AQP1 (NP_000376). Gray columns indicate the two NPA motifs and the cysteine residue that are responsible for mercury inhibition of water transport. The two horizontal gray bars indicate the residues against which the two antibodies used in the present study were raised (2, 28). An asterisk (*) indicates that the amino acid residues are identical in all sequences in the alignment: a colon (:) indicates conserved substitutions: and a period (.) indicates semiconserved substitutions.