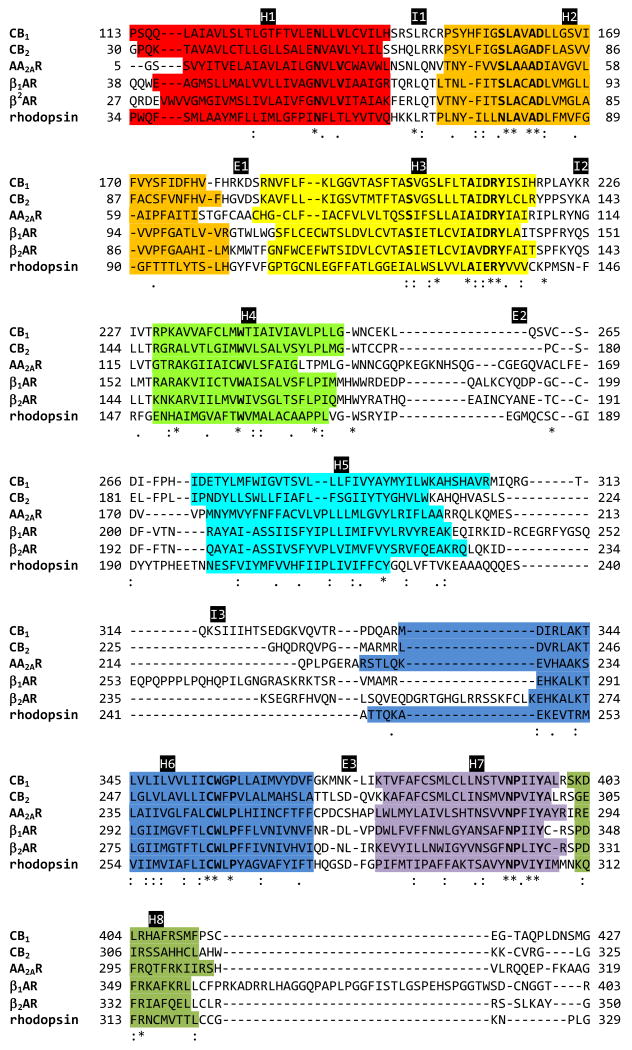
Fig. (1).
Sequence alignment by T-COFFEE (V7.71, mode: expresso) (http://www.tcoffee.org) [211] of CB1 and CB2 with the GPCRs whose structures have been determined by the X-ray crystallography, including AA2AR (PDB code: 3EML) [42], β1AR (PDB code: 2VT4) [38], β2AR (PDB code: 2RH1) [40], and rhodopsin (PDB code: 1U19) [46]. The TM helical boundaries for the CB1 and CB2 receptors are from the respective homology models [136,182], while the TM helical boundaries for AA2AR, β1AR, β2AR, and rhodopsin are from the respective X-ray structures. Conservancy of the aligned sequence by CLUSTALW (http://www.ebi.ac.uk/Tools/clustalw/) [212] is represented by consensus symbols: “*” for identical residues; “:” for conserved substitutions; and “.” for semi-conserved substitutions. Highly conserved residues in the rhodopsin family of GPCRs reported by Baldwin et al (1997) are in bold. Color codes for TM helices: H1 (in red); H2 (in orange); H3 (in yellow); H4 (in green); H5 (in cyan); H6 (in blue); H7 (in purple); and H8 (in dark green).