Abstract
Objectives
The National Cancer Institute (NCI) has developed the Common Data Elements (CDE) to serve as a controlled vocabulary of data descriptors for cancer research, to facilitate data interchange and inter-operability between cancer research centers. We evaluated CDE’s structure to see whether it could represent the elements necessary to support its intended purpose, and whether it could prevent errors and inconsistencies from being accidentally introduced. We also performed automated checks for certain types of content errors that provided a rough measure of curation quality.
Methods
Evaluation was performed on CDE content downloaded via the NCI’s CDE Browser, and transformed into relational database form. Evaluation was performed under three categories: 1) compatibility with the ISO/IEC 11179 metadata model, on which CDE structure is based, 2) features necessary for controlled vocabulary support, and 3) support for a stated NCI goal, set up of data collection forms for cancer research.
Results
Various limitations were identified both with respect to content (inconsistency, insufficient definition of elements, redundancy) as well as structure – particularly the need for term and relationship support, as well as the need for metadata supporting the explicit representation of electronic forms that utilize sets of common data elements.
Conclusions
While there are numerous positive aspects to the CDE effort, there is considerable opportunity for improvement. Our recommendations include review of existing content by diverse experts in the cancer community; integration with the NCI thesaurus to take advantage of the latter’s links to nationally used controlled vocabularies, and various schema enhancements required for electronic form support.
Keywords: Common Data Elements, Cancer Research, Metadata Registries, ISO/IEC 11179, Cancer Bioinformatics Grid
1 Introduction
The National Cancer Institute’s Cancer Bioinformatics Grid (CaBIG, http://cabig.nci.nih.gov)(1) comprises a network of individuals from NCI-supported cancer centers, NCI personnel and NCI-affiliated contractors, who are working towards the creation of standards for cancer-related informatics, and the eventual creation of interoperable software modules supporting those standards. The modules will serve various purposes, from exchange of research data, conduct of clinical trials, financial, billing and other administrative tasks, adverse event reporting, and so on.
The interoperation will be based on common metadata standards. (The term “metadata” – data that describe and define other data” (2) – is used in both the singular and plural.) Among the various forms of metadata are controlled vocabularies, whose role in biomedical standardization efforts is well known. Examples of biomedical controlled vocabularies are the Medical Subject Headings (MeSH) (3) Logical Observations, Identifiers, Names and Codes (LOINC) (4), the Systematic Nomenclature of Medicine (SNOMED) (5) and the Gene Ontology (GO) (6). The National Library of Medicine (NLM)’s Unified Medical Language System (UMLS)(7) is a compendium of numerous existing biomedical vocabularies, including all those just mentioned.
The NCI has developed two controlled vocabularies. One of these, the NCI Thesaurus (8, 9), is incorporated into UMLS. The other, the Common Data Elements (CDE) is not, and its contents are consequently less well known. The NCI Center for Bioinformatics (NCICB) describes the purpose of CDE as follows(10):
“One of the problems confronting the biomedical data management community is the panoply of ways that similar or identical concepts are described. Such inconsistency in data descriptors (metadata) makes it nearly impossible to aggregate and manage even modest-sized data sets in order to be able to ask basic questions. The NCI, together with partners in the research community, develops common data elements (CDEs) that are used as metadata descriptors for NCI-sponsored research…CDEs are descriptors of data – metadata – that are used to set up data collection forms for cancer research studies.”
Various NCI-sponsored groups conduct clinical research that generates significant amounts of data. The parameters that are recorded relate to clinical and laboratory findings as well as items in standardized questionnaires. Just as individual items in an laboratory data stream using the Health Level 7 (HL7) communications protocol (www.hl7.org) are tagged with LOINC identifiers for the corresponding lab parameters, the idea is that eventually CDEs could act similarly as a foundation for cancer research data interchange. Most centers use a variety of clinical study data management systems (CSDMSs) for electronic data collection during conduct of clinical research. If such software is initialized with CDE content, then, when electronic forms are set up, mapping of the form elements/questions to CDEs would greatly simplify the interchange of data collected at different cancer centers. An additional hope is that existing definitions of standard cancer research forms may be reused in their entirety instead of having to be redefined by each group.
The CDE has some attributes of a controlled terminology, in the sense that its contents, like LOINC identifiers or SNOMED-CT concepts, will be utilized far beyond their site of origination to support semantic mapping between electronic systems whose use is related to cancer research or treatment. An internal evaluation of CDE from the terminology aspect has been previously performed by the Chute group at Mayo Clinic; its conclusions are reported very briefly on the group’s Web site.(11). We analyzed functions and structure to determine its fitness for its intended purpose.
2 Background: CDE Design Principles
The CDE, which has been in continuous development for at least four years, was originally intended to be a standard nomenclature for the reporting of Phase 3 cancer clinical trials data (12). NCICB stores the CDE in a relational database called caDSR (Cancer Data Standards Repository), whose design is influenced by the ISO/IEC 11179 standard for descriptive metadata (13) (ISO=International Standards Organization; IEC=International Electrotechnical Commision). The documents describing ISO/IEC 11179 prescribe a conceptual model rather than an actual physical implementation, even though they include several Unified Modeling Language (UML) (14) class diagrams. These diagrams, an extended form of the Entity-Relationship diagrams used to model database schemas, also incorporate referential integrity constraints between the various components. (An example of a referential integrity constraint for an outpatient clinic database is: if a new visit is recorded for a patient, the “physician visited” must first exist in the database.) Referential integrity is so important that modern database engines allow it to be specified “declaratively”, through a concise phrase in the schema definition language, as opposed to having to write code.
In ISO/IEC 11179 terminology, a Data Element is the fundamental unit of data that an organization disseminates. A data element is based on a Data Element Concept (the abstract unit of knowledge that it represents) and a representation. Aspects of representation include unit of measure, and Value Domain. A value domain (a set of permissible/valid values for the element) is defined by information such as data type (e.g., number, character, date), maximum and minimum permissible values, maximum and minimum permissible length (e.g., number of characters), number of decimal places, and if applicable, an enumerated list of values (typically codes accompanied by descriptive phrases).
One composes a data element by combining a concept with a value domain. In some cases, only one value domain is meaningful for a given concept. In other cases, however, an abstract concept may be described in more than one way, e.g., quantitatively (as numbers in a specified unit), qualitatively (absent, mild, moderate, severe) or comparatively with a reference (e.g., above normal, within normal limits, or below normal), and each type of description calls for a different value domain. Obviously, a single value domain may often apply to multiple data elements. Some value domains occur so commonly that they may be treated as special data types: a well-known example is the Boolean data type, which consists of the enumeration (True/Yes, False/No).
Clear guidelines are provided for composing the names and definitions of data elements from the names of the concepts and the value domains. Related concepts may be grouped into Classes, but the standard leaves the details of this issue unspecified.
3 Methods
3.1 CDE Content and Structure
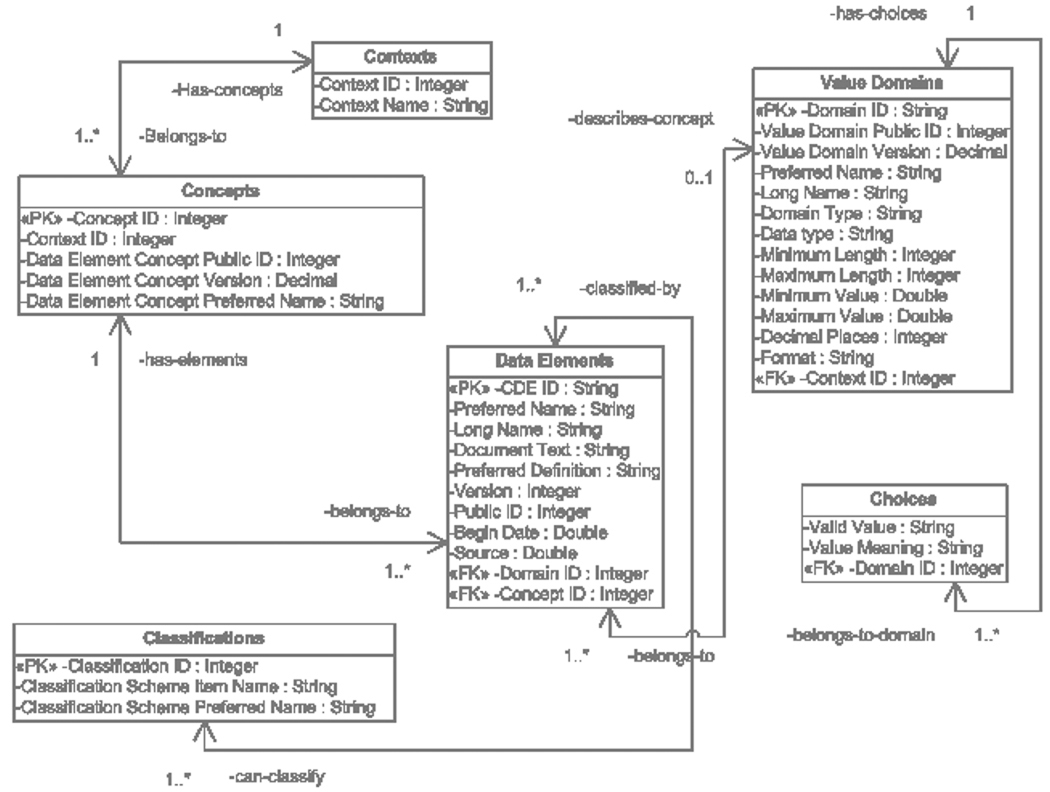
NCICB does not provide direct access to the caDSR schema or an ftp-able version of the contents plus schema definition. We therefore accessed CDE content via the CDE browser (15), which performs a live query of the database. The downloaded contents are in the form of XML or a delimited file containing 57 columns. This file is the result of a join of around nine relational tables, and is consequently highly redundant in content. With some programming effort and the help of the CDE technical documentation diagrams, we reconstructed a semantically equivalent copy of the original schema. The UML class diagram illustrating the schema, and the relationships between tables, is illustrated in fig. 1. (A Microsoft Access database containing the schema and complete CDE contents as of Sept. 1, 2004 can be downloaded from our ftp site as ftp://custard.med.yale.edu/others/cde.zip.)
Figure 1.
A Unified Modeling Language (UML) Class Diagram describing CDE content. The key classes tables from the perspective of element use are Concepts, Value Domains, Data Elements and Choices. The classes are implemented as relational tables: in each class, the symbols ≪PK≫ and ≪FK≫ indicate primary and foreign keys, respectively.
To understand this structure, it helps to emphasize that the ISO/IEC 11179 model is not intended to represent metadata with sufficient richness to address every potential use for it. This model is concerned specifically with the structure of “metadata registries” – official repositories of metadata gathered from different sources. Many aspects of the CDE schema are concerned with issues of provenance (origin, attribution)– which source created or is responsible for a particular element, what the current version of a particular element is, and what it is designated as in the source, etc. These aspects have been elided in the figure by shortening or omitting the details of certain tables.
The four most important tables in fig. 1 are Concepts, Value Domains, Data Elements and Choices. As stated earlier, a data element is logically a combination of a concept with a value domain, and therefore the Data Elements table acts as a “bridge” between the other two. For value domains that comprise a list of enumerated or ordinal items, the individual items are stored in Choices. Both a concept and a value domain may be derived from a particular source (in UMLS terminology), which in ISO/IEC 11179 is called a Context. Each context has an administrator with the authority to manage and edit the CDEs that the context “owns”. Examples of Contexts are individual NCI divisions such as CTEP (Cancer Therapy Evaluation Program) or the SPORE research consortia (Specialized Programs of Research Excellence). A data element may be indexed by one or more keywords: the table Classifications records these keywords. Finally, there may be documentation records associated with an element, as well as entries (“Designations”) that denote how the data element is recorded in the original source: this last is roughly equivalent in function to the Source Abbreviation field in UMLS.
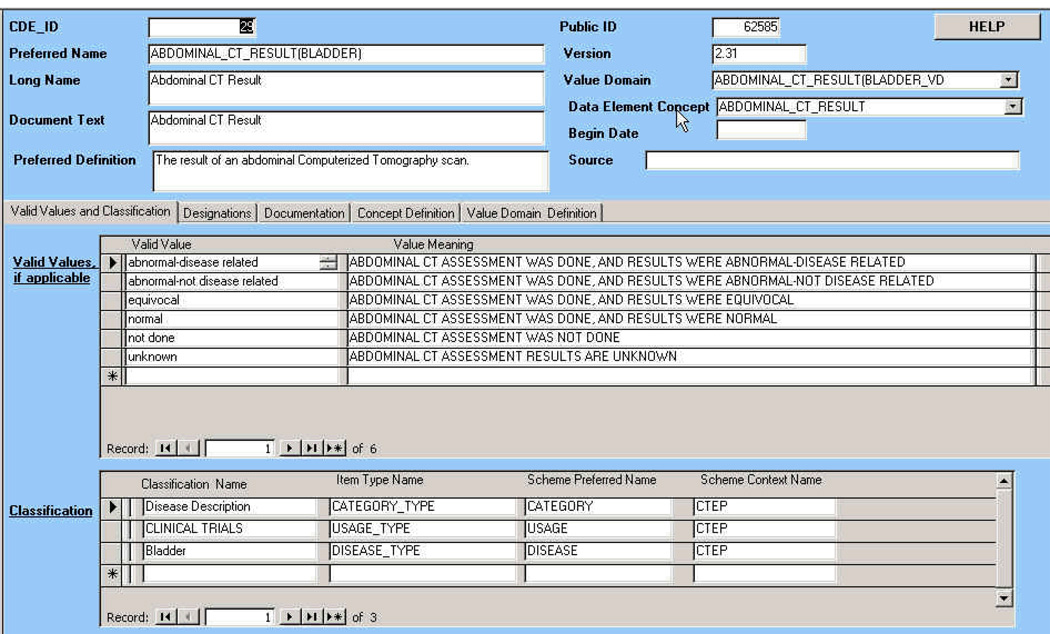
Fig. 2 is a screen-shot of a form within the Microsoft Access application, showing the details of an individual data element (the summary result of an abdominal CT scan used to assess bladder disease). Associated data in the Concepts, Value Domains, Choices and Classification tables is also displayed in the same form.
Fig. 2.
Details of an individual Common Data Element (the summary result of an abdominal CT scan to assess disease). The Value Domain and Concept that this CDE.belongs to are shown on the top right, while the individual Choices in the value domain and the various Classification categories that apply to this element are shown in lists on the lower part of the screen. Note that the names of the Value Domain and the Concept associated with the Data Element are highly similar to that of the element itself (using the string “ABDOMINAL_CT_RESULT”), indicating that they have possibly been algorithmically generated. This follows the requirements that every data element must be associated with a concept as well as a domain: both of these must be created accordingly if they did not exist in the database previously.
3.2 Evaluation Categories and Approach to Evaluation
We evaluated the caDSR and its content (the CDEs and related tables described in fig.1) from several aspects:
Compatibility with the ISO/IEC logical metadata model, on which it is based.
Support for features that are considered standard or highly desirable for controlled vocabularies in general.
Its intended purpose – support for electronic data entry form setup and content validation.
Each of these aspects was evaluated from several perspectives:
-
Structure: Does the existing structure (i.e., the database schema) provide a means to represent an essential or highly desirable feature to support an intended purpose? Identification of structural differences (desired vs. actual) is qualitative, but can have fairly direct quantitative consequences: for example, if the structure to support recording synonyms of concepts is not present, one can infer that synonymy coverage is zero. In previous issues of this journal, Cimino has used this technique to identify desiderata for controlled vocabularies, and then determined the extent to which existing vocabularies have the structure to support these desiderata(16). Cimino has also characterized the structures necessary to support of change in vocabularies (17).
An evaluation of structure was used for ISO 11179 compatibility, support of controlled vocabulary features, and support of electronic form representation. The “technique” of structure evaluation involves comparison of the CDE structure with schemas/class diagrams that are known to serve a specific purpose, i.e., the ISO 11179 class diagrams, the SNOMED schema (for controlled vocabularies) and our own TrialDB (for E-form representation).
-
Constraints: Does the existing structure have means to prevent inconsistencies or errors from being accidentally introduced into the content? An important family of constraints deals with referential integrity, described earlier: when not enforced, errors can creep into the content.
Evaluation from the constraint perspective was used to test aspects of adherence to ISO 11179. We identified necessary constraints from the ISO 11179 class diagrams, and then composed, for each constraint, a SQL query to quantify the records that violated that constraint. (A similar query-based approach was taken by Olivier Bodenreider of the NLM in detecting and quantifying the number of circular hierarchical relationships in the UMLS (18))
Content: Given that the CaDSR content is intended to serve as a reference terminology intended to be accessed nationally and internationally by cancer centers, to what extent is its content “controlled”? Content evaluation is human-intensive and is necessarily sample-based: an example of content evaluation is Hersh et al’s comparative evaluation of SNOMED and UMLS for mapping of phrases encountered in clinical text to vocabulary concepts (19). We detected the presence of certain types of curation errors that could provide a rough indication of curation rigor. We quantified redundancy (content duplication) through SQL queries that look for duplicated strings in individual tables (e.g., in definitions).
Completeness of Evaluation: Because CDE has few tables and a simple structure, a structural evaluation can be complete. Similarly, evaluations based on the running of queries that test specific constraints or duplications can also be complete, because they return all rows that fail the desired criterion. However, certain kinds of content errors, such as errors in semantics, can only be identified by visual inspection of individual rows of data and application of domain knowledge. Because of the size of CDE, one cannot quantify these errors precisely without intensive curation. Our evaluation from the latter aspects cannot be considered “complete”: however, it is still important to report their presence where they are encountered in the course of another aspect of the evaluation.
The results are now described under these headings.
4 Results of Evaluation
4.1 caDSR’s Compatibility with ISO/IEC 11179
While caDSR design follows the logical model for ISO/IEC 11179, it departs from this model in several ways, as reflected in both structure as well as content. This divergence can be problematic. We now explain with examples.
-
Referential Integrity Issues: Though a data element is supposed to be derived from a concept-domain pair, a small percentage of data elements (about 150 out of 11,400 records, or 1.32%) lack both a parent concept and a parent value domain. Some of these entries are spurious terms, as indicated by the element name containing strings like “test” or ‘fdgfg”.
By the ISO/IEC 11179 definition, every item in the Choices table must belong to a particular domain (i.e., the domain ID field in this table must be non-empty and valid). 520 out of 35,300 choice records (1.47%) are not associated with a domain. This is why the schema diagram of fig. 1 has a cyclical relationship between the tables Choices, Data Elements and Value Domains. Ideally the link between Choices and Data Elements would not be necessary because it can be inferred.
While the above errors can be corrected by manual curation, their permanent prevention requires creating additional database constraints within caDSR.
-
Content Duplication and Separation of Function: Some content in the CDE exhibits redundancy. According to the ISO/IEC 11179 model, a concept-value domain pair uniquely defines a data element semantically. Within caDSR, however, there are numerous data elements (1963 records out of 11,400 CDEs, or 17.22%) where the concept ID-value domain ID pair is not unique. An example is the concept “Abominal CT assessment date”, to which a single domain (the range of all valid dates) applies. There are two data elements, both with the preferred definition “The date an abdominal computerized tomography scan is assessed.” Both data elements come from the CTEP source. One of these is called “ABDOMINAL_CT_ASSESSM(PROSTATE)”, and the other is “ABDOMINAL_CT_ASSESSME(BLADDER)”. It appears that abdominal CT is an assessment used in two different CTEP cancer evaluation forms. Another instance is the concept of the State Code part of the postal address, for which there are five data elements, “ADDRESS_STATE_CD”, “ADDRESS_STATE_CODE(LUNG)”, “…(BLADDER)”, ”…(PROSTATE)” and “…(BREAST)”. All these elements have identical descriptions and attribute properties.
If it is considered important to record all instances where a particular logical data element is used, this should be recorded separately rather than creating multiple semantically identical elements. Proliferation of data elements in this way thwarts their intended purpose of reuse by cancer centers when developing their own electronic data entry forms. For example, suppose a protocol for evaluation of metastatic liver cancer required an abdominal CT, how would one readily decide which of the elements “ABDOMINAL_CT_ASSESSM(PROSTATE)” or “…(BLADDER)” was appropriate? Obviously, on closer inspection, one would realize that either could be used. Much later, however, when the form was deployed in production and the metadata defining the form (or a user-friendly version of a tagged data stream) was inspected, its interpretation would be confusing. What does a prostate-related parameter have to do with liver cancer?
-
Content Descriptions: The ISO/IEC 11179 model requires that metadata curators make a significant effort to provide clear and unambiguous definitions for individual metadata items, since these are intended for human inspection. The level of annotation of caDSR content is currently highly variable. In general, common data elements tend to be better annotated than their concepts from which they derive. In fact, while the Data Elements table has a column called “Preferred Definition”, in addition to “Preferred Name” and “Long Name” columns, both the Concepts and Value Domains tables lack a preferred definition column, so that in effect there is permission to leave these entities inadequately annotated. Consequently, there are concepts and value domains whose purpose cannot be inferred by inspecting them in isolation.
An example is the concept with the long name “Tumor Description” (preferred name “TUMOR_DESC”). This concept has 68 “child” data elements: some elements are related to tumor staging by “tumor-node-metastasis” (T-N-M) criteria for various organ systems by clinical or pathologic criteria, while other elements are related to tumor grading by histology for various organ systems. One can only infer this concept’s purpose by inspecting these “child” elements. Value domains whose intent cannot be inferred at all are the three with the shared preferred name “UNKNOWN_CVD”, which can take 0–1 characters. These three domains apply to about 432 data elements. The existence of three “Unknown_CVD” domains rather than one also appears to be a case of redundancy: all three have identical property values.
Domain Definition Errors: Several domain definitions are incorrect given the semantics of particular data elements. An example is the data element “HMT_LYMP_LAB_PTG_VAL” (the lymphocyte percentage in the differential white cell count). The domain for this element is defined as having minimum length zero, maximum length five and zero decimal places. The minimum and maximum permissible values are left unspecified. In reality, we know from knowledge of the WBC differential that the minimum and maximum values must be zero and 100 respectively, while the maximum length cannot exceed 3 digits (=100).
4.2 Controlled Vocabulary/Ontology Features
4.2.1 Support of Inter-Concept Relationships
Requirement: Controlled vocabularies should provide means of arranging related concepts of varying granularity in a hierarchy or network. Recording hierarchical and non-hierarchical inter-concept relationships, as in SNOMED and UMLS, supports navigational browsing, facilitating understanding of the vocabulary’s coverage, and helps to identify potential redundancies.
The concepts within caDSR range from finely granular concepts like “line 1 of the street address”, to concepts like “Hematology Lab”, which includes numerous diverse data elements such as erythrocyte sedimentation rate, prothrombin time and the various components of the differential white blood cell count. However, caDSR lacks such a hierarchy; all concepts exist at a single level, with no means of inter-relating them. The Mayo document cited earlier points out that the absence of semantic or syntactic linkage of shared concepts makes it difficult to algorithmically recognize related.concepts (11)
Another consequence of the lack of this desideratum is content redundancy. For example, The “Hematology Lab” concept has a data element with the preferred name HMT_NEUT_LAB_PTG_VAL and the definition “peripheral blood neutrophils percentage”. On the other hand, neutrophil cell percentage is also a concept in its own right. The data element that is the single child of this concept, however, is different from the one just mentioned. It has the preferred name “LAB_HEME_NEUTROPHILS_CELL_*” and the same preferred definition, except that it begins with a capital P. Similar redundancies occur for other parts of the differential, such as monocytes, promyelocytes, etc., as well as the absolute counts of these cell types. This situation represents one of unrecognized synonyms, since the underlying semantics of the two data elements are identical.
4.2.2 Support of Synonymy
Requirements: A controlled vocabulary must support representation of alternative synonymous forms (terms) for the same underlying concept. The clinical domain, for example, has both Anglo-Saxon vs. Greco-Latin equivalents for the same concepts, e.g., vomiting vs. emesis. Terms, or the key phrases that they contain, provide a means of query expansion, in that the same concept can be located through different search terms.
The caDSR lacks a “synonyms/terms” table. Concepts are only classified by keywords and grouped by the source they came from. Consequently, searching is less robust.
4.3 Support for Electronic Form Definition
Requirements: In order to support generation of robust electronic data forms from individual data elements, it is necessary to record information that inter-relates these elements, such as:
The order in which the elements should be presented to the user. In psychiatric research, for example, changing the order of questions on a form can alter the form’s meaning and interpretation (20). (Psychiatry forms used in cancer research include the Center for Epidemiologic Studies Depression Scale (CES-D), which has been used for evaluation of reactive depression following disfiguring surgery for head and neck cancers (21, 22), The order of questions in the CES-D is required to remain fixed.)
Rules/Formulas for Computation of Certain Elements based on the values of other elements that precede the computed element in the form. Body surface area, for example, is computed as a function of height and weight by the Dubois formula, BSA in meters2 = 0.20247*(height in meters^0.725) * (weight in Kg ^0.425). An important issue here is the choice of programming language used to specify the formula: the programming languages C, Perl and JavaScript do not have a built-in exponentiation operator.
Dynamic enabling or disabling of certain elements based on responses to preceding elements (skip logic): This helps to ensure content validity. For example, a set of questions regarding diabetes is inapplicable if the patient or the patient’s relatives do not have this condition.
Cross-element validation through arbitrary complex rules: For example, the sum of the individual differential WBC components should be exactly 100.
The papers of van Ginneken (23), which addresses requirements for structured data entry, and Nadkarni et al (24) which deals with E-form generation for a clinical trials database, address this theme in greater detail: the software described in both papers is available as open-source. More than a dozen relational tables are required to capture the requisite information, starting with the representation of a form itself. (The documentation and code for our open-source clinical study data management system, TrialDB (25, 26), are available at ftp://custard.med.yale.edu).
The CDE schema currently lacks the structures necessary to record any of the information necessary for form definition as delineated above. Meeting this goal is realistic, but the details of how it may be achieved are beyond the scope of this paper.
5 Discussion
To recapitulate the results of the evaluation:
The CaDSR structure differs from ISO 11179 in being less rigorous: many constraints and integrity checks defined in the standard are not implemented. This results in duplication of content, and certain types of curation errors that could have been prevented were these checks in place.
While the CaDSR content (the CDE) is intended to serve the purposes of a controlled terminology, the basic structures to support controlled terminologies, namely representation of concept synonyms and inter-concept relationships, are not present. The consequences are inability to recognize related concepts as related, as well as content redundancy.
The structures necessary to meet the representation of Electronic data entry forms, a stated goal of the CaDSR, are lacking.
As indicated by detection of several types of curation errors, the rigor of content curation falls short of the level expected of a standard intended for national deployment.
Despite the problems identified in the evaluation, there are certain positive aspects to CDE initiative, notably the decision to utilize the existing ISO/IEC 11179 standard model has raised general awareness of this standard (previously confined mostly to the information-technology world) in the biomedical informatics community. Several of this model’s aspects – such as the definition of value domains –benefit all controlled biomedical vocabulary efforts, by addressing several issues that existing vocabularies currently tend to deal with in an ad hoc fashion.
Mere adoption of a standard model, however, does not suffice. The current version of ISO 11179 was devised for support of registries of descriptive metadata, not for the considerably more complex issues that CDE tries to address. Several HL7-affiliated investigators have recognized this limitation and are working towards augmenting ISO 11179 for ontology support (notably Harold Solbrig of Mayo Clinic, whose previous work on ISO 11179 is described in (27)) but it may take some time before these efforts yield a revised standard data model.
To their credit, caBIG affiliated individuals have begun to establish well-defined processes for new data elements suggested for incorporation into CDE as standard elements (28), including human expert review of the existing CDE content to check for semantic identity with an existing element. Much existing (legacy) content, however, has attained a “standard” status as a consequence of less rigorous review processes. The recommendations below deal with how to fixing CDE structure and content to support its various intended roles.
6 Recommendations
We state our recommendations under the three broad categories of evaluation stated earlier.
6.1 ISO/IEC 11179 Compatibility and Content
NCICB needs to ensure that CDE content adheres to the standard’s intentions of clear, correct and unambiguous definitions and descriptions. This task requires curatorial input from cancer/clinical content experts as well as those with experience in development of biomedical thesauri. Relatively few individuals outside NCI have had an opportunity to inspect CDE content in its entirety. It is desirable for NCICB to emulate the example of UMLS, whose contents are made available as a set of delimited text files whose contents can readily be massaged and imported into relational tables. The present hurdle of requiring those who wish to inspect CDE content in bulk to parse a complex-structured and highly content-redundant XML stream, results in unnecessarily duplicated effort at individual cancer centers.
It is desirable to follow the example of UMLS and explicitly support preferred definitions for both concepts and domains. The most useful source vocabularies that feed into UMLS (notably the NLM’s Medical Subject Headings) record concept definitions.
6.2 Support for Controlled Vocabulary Features
Standard structures to support the minimum requirements of controlled vocabularies–synonyms and relationships – should be incorporated. This will also facilitate mapping of CDE content to standard sources such as the UMLS, and leveraging UMLS content in turn to link to its constituent vocabularies such as LOINC and SNOMED. One of the efforts that is part of the CaBIG initiative is the creation of an information architecture that faithfully follows the HL7 version 3 draft standard (29): such mapping will facilitate interpretation of the semantics of data streams that contain CDEs as attributes.
The NCI thesaurus already has mapping to UMLS in place, but the CDE and NCI thesaurus efforts currently appear to be operating and managed more or less independently of each other. Integration of CDE content into the NCI thesaurus should be a high priority. The use of “terminology services” tools such as developed by the Mayo group (30–32) should help greatly in such an integration effort.
CDE will possibly need to borrow eventually from SNOMED-CT to incorporate a mechanism for support of description logics. The research of Hahn and Schulz (33, 34) and the OpenGALEN project (35, 36) has shown that controlled terminologies often need to be augmented by mechanisms for such knowledge representation. SNOMED-CT currently uses an XML representation to support composition of complex concepts from more atomic ones, and CDE will possibly need to use a similar approach.
Given the modest size of CDE, the tasks of enforcing ISO 11179 compliance and controlled vocabulary features are tractable
6.3 E-form Support
To meet the goal of supporting computable electronic data collection forms, the caDSR schema requires major extensions. The complexity of this task, however, cannot be underestimated. In our own experience in maintaining a clinical trials data management system over more than seven years, this schema component has evolved continually to meet user demands. For example, we have now added metadata schema (and generic code) to support dynamic E-form generation in an arbitrary number of languages (e.g., English, Spanish, German). Such a feature is useful in clinical studies that are conducted internationally, because it allows a single Web site to serve pages in multiple languages without creating multiple bodies of code or multiple database schemas. At the metadata schema level, such support involves allowing multiple display captions for the same data element (and for each value in an enumerated value domain), one for each target language. Ideas from the UMLS, which records synonymous terms for the same concepts in different languages, can be profitably borrowed.
We believe that the goals of full ISO/IEC 11179 compatibility and schema infrastructure for controlled terminologies are achievable with relatively modest resources, while E-form support should be postponed until these goals are met.
7 Conclusions
The CDE is a critical linchpin in the highly desirable goal of inter-operability and data sharing for cancer research. This evaluation is intended to assist the cancer informatics community in identifying ways of improving the CDE.
Acknowledgements
NIH grants U01 CA78266, U01 ES10867, R01 LM06843 and K23 RR16042, institutional funds from Yale University School of Medicine, and a contract from the National Cancer Institute for support of participation in the Cancer Bioinformatics Grid (caBIG)
Bibliography
- 1.National Cancer Institute. [Last accessed: 11/25/04];Cancer Bioinformatics Grid. 2004 http://cabig.nci.nih.gov.
- 2.Marco D. Building and Managing the Metadata Repository. New York: Wiley; 2000. [Google Scholar]
- 3.National Library of Medicine. [Last accessed: 11/25/04];Medical Subject Headings- Home Page. 2004 www.nlm.nih.gov/mesh/meshhome.html.
- 4.Regenstrief Institute. [Last accessed: 7/8/02];LOINC home page. 2002 http://www.regenstrief.org/loinc/
- 5.College of American Pathologists. [Last accessed: 10/2/02];SNOMED Clinical Terms (SNOMED CT) 2002 www.snomed.org.
- 6.Gene Ontology Consortium. [Last accessed: 11/26/04];An Introduction to Gene Ontology. 2004 http://www.geneontology.org/GO.doc.html.
- 7.Lindberg DAB, Humphreys BL, McCray AT. The Unified Medical Language System. Meth. Inform. Med. 1993;32:281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Cancer Institute. [Last accessed: 11/25/04];Terminology Resources: NCI Thesaurus and Enterprise Vocabulary Services (EVS) 2004 http://www.nci.nih.gov/cancertopics/terminologyresources.
- 9.De Coronado S, Haber MW, Sioutos N, Tuttle MS, Wright LW. NCI Thesaurus: Using Science-based Terminology to Integrate Cancer Research Results. Medinfo. 2004;2004:33–37. [PubMed] [Google Scholar]
- 10.National Cancer Institute. [Last accessed: 11/25/04];Cancer Data Standards Repository (caDSR) 2004 http://ncicb.nci.nih.gov/core/caDSR.
- 11.Mayo Clinic Biomedical Informatics Group. [Last accessed: 12/4/04];Semantic Structures for Patient Data Retrieval. 2004 http://mayoresearch.mayo.edu/mayo/research/bmi/grant_sspdr_suppl_2_full.cfm.
- 12.Meadows B, Abrams J, Christian M, Silva J, Pifer C, Valmonte C, et al. The Common Data Elements Dictionary- A Standard Nomenclature for the Reporting of Phase 3 Cancer Clinical Trials Data; 14th IEEE Symposium on Computer-Based Medical Systems; 2001; Bethesda, MD. Los Alamitos, CA: IEEE Press; 2001. [Google Scholar]
- 13.International Standards Organization. [Last accessed: 11/05/04];ISO/IEC 111179, Information Technology - Metadata Registries. 2004 http://metadata-stds.org/11179/
- 14.Booch G, Rumbaugh J, Jacobson I. The Unified Modeling Language User Guide. Reading, MA: Addison-Wesley; 1998. [Google Scholar]
- 15.National Cancer Institute. [Last accessed: 11/25/04];CDE Browser. 2004 http://cdebrowser.nci.nih.gov/CDEBrowser/
- 16.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 17.Cimino JJ. Formal descriptions and adaptive mechanisms for changes in controlled medical vocabularies. Methods Inf Med. 1996;35(3):202–210. [PubMed] [Google Scholar]
- 18.Bodenreider O. Proceedings of the AMIA Fall Symposium; 2001. Washington DC: Hanley & Belfus; 2001. Circular Hierarchical Relationships in the UMLS: Etiology, Diagnosis, Treatment, Complications and Prevention; pp. 57–61. [PMC free article] [PubMed] [Google Scholar]
- 19.Hersh WR, Campbell EH, Evans DA, Brownlow ND. Proceedings/AMIA Annual Fall Symposium; 1996. 1996. Empirical, automated vocabulary discovery using large text corpora and advanced natural language processing tools; pp. 159–163. [PMC free article] [PubMed] [Google Scholar]
- 20.White TM, Hauan MJ. Extending the LOINC Conceptual Schema to Support Standardized Assessment Instruments. J Am Med Inform Assoc. 2002;9:586–599. doi: 10.1197/jamia.M1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Graeff A, de Leeuw JR, Ros WJ, Hordijk GJ, Blijham GH, Winnubst JA. Sociodemographic factors and quality of life as prognostic indicators in head and neck cancer. Eur J Cancer. 2001;37(3):332–339. doi: 10.1016/s0959-8049(00)00385-3. [DOI] [PubMed] [Google Scholar]
- 22.Katz MR, Kopek N, Waldron J, Devins GM, Tomlinson G. Screening for depression in head and neck cancer. Psychooncology. 2004;13(4):269–280. doi: 10.1002/pon.734. [DOI] [PubMed] [Google Scholar]
- 23.van Ginneken AM. Considerations for the representation of meta-data for the support of structured data entry. Methods Inf Med. 2003;42(3):226–235. [PubMed] [Google Scholar]
- 24.Nadkarni PM, Brandt CA, Marenco L. WebEAV: Automatic Metadata-driven Generation of Web Interfaces to Entity-Attribute-Value Databases. Journal of the American Medical Informatics Association. 2000;7(7):343–356. doi: 10.1136/jamia.2000.0070343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadkarni PM, Brandt C, Frawley S, Sayward F, Einbinder R, Zelterman D, et al. Managing attribute-value clinical trials data using the ACT/DB client-server database system. Journal of the American Medical Informatics Association. 1998;5(2):139–151. doi: 10.1136/jamia.1998.0050139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandt C, Nadkarni P, Marenco L, Karras B, Lu C, Schacter L, et al. Reengineering a database for clinical trials management: lessons for system architects. Controlled Clinical Trials. 2000;21(5):440–461. doi: 10.1016/s0197-2456(00)00070-2. [DOI] [PubMed] [Google Scholar]
- 27.Solbrig H. Metadata and the Reintegration of Clinical Information: ISO 1179. MD Computing. 2000 May–June;:25–28. [PubMed] [Google Scholar]
- 28.Curtis T. [Last accessed: 12/4/04];Common Data Elements (CDEs) Harmonization. 2004 http://cabig.nci.nih.gov/workspaces/VCDE/Documents/Useful_Presentations/CDEs/caBIGIntegratedCancerResearchHarmonization%20082404FINAL.pdf.
- 29.Hammond W. [Last accessed: 4/14/04];Introduction to HL7. 2003 www.hl7.cz/doc/EasternEuropeTutorial.ppt.
- 30.Solbrig HR, Chute CG. Terminology Access Methods Leveraging LDAP Resources. Medinfo. 2004;2004:545–549. [PubMed] [Google Scholar]
- 31.Savova GK, Becker D, Harris M, Chute CG. Combining Rule-based Methods and Latent Semantic Analysis for Ontology Structure Construction. Medinfo. 2004;2004:1848. (CD) [Google Scholar]
- 32.Solbrig HR, Armbrust DC, Chute CG. The Open Terminology Services (OTS) project. AMIA Annu Symp Proc. 2003:1011. [PMC free article] [PubMed] [Google Scholar]
- 33.Schulz S, Romacker M, Hahn U. Part-whole reasoning in medical ontologies revisited--introducing SEP triplets into classification-based description logics. Proc AMIA Symp. 1998:830–834. [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn U, Schulz S. Towards a broad-coverage biomedical ontology based on description logics. Pac Symp Biocomput. 2003:577–588. doi: 10.1142/9789812776303_0054. [DOI] [PubMed] [Google Scholar]
- 35.Rector AL, Rogers JE, Zanstra PE, Van Der Haring E. OpenGALEN: open source medical terminology and tools. AMIA Annu Symp Proc. 2003:982. [PMC free article] [PubMed] [Google Scholar]
- 36.Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL concept modelling language for medical terminology. Artif Intell Med. 1997;9(2):139–171. doi: 10.1016/s0933-3657(96)00369-7. [DOI] [PubMed] [Google Scholar]