Abstract
African-Americans have higher rates of kidney disease than European-Americans. Here we show that in African-Americans, focal segmental glomerulosclerosis (FSGS) and hypertension-attributed end-stage kidney disease (H-ESKD) are associated with two independent sequence variants in the APOL1 gene on chromosome 22 [FSGS odds ratio = 10.5 (95% CI 6.0–18.4); H-ESKD odds ratio = 7.3 (95% CI 5.6–9.5)]. The two APOL1 variants are common in African chromosomes but absent from European chromosomes and both reside within haplotypes that harbor signatures of positive selection. ApoL1 is a serum factor that lyses trypanosomes. In vitro assays revealed that only the kidney disease-associated ApoL1 variants lysed Trypanosoma brucei rhodesiense. We speculate that evolution of a critical survival factor in Africa may have contributed to the high rates of renal disease in African-Americans.
African-Americans suffer from kidney failure at high rates compared with individuals without recent African ancestry(1–3). Genetic variation at a locus in or near the MYH9 gene on chromosome 22 has been associated with the increased susceptibility to focal segmental glomerulosclerosis (FSGS), HIV-associated nephropathy, and hypertension-attributed end-stage kidney disease (H-ESKD) observed in African-Americans(4, 5), but thus far causal mutations in MYH9 have not been identified(6–8).
Previous genome-wide analyses have shown a strong signal of natural selection in the region containing the MYH9 and APOL1 genes (iHS data available at http://hgdp.uchicago.edu/)(9–13). This observation led us to hypothesize that the kidney disease risk alleles might be located in a larger interval than originally thought(4, 5). The longer patterns of linkage disequilibrium (LD) associated with variants undergoing selection suggests that a positively selected risk variant could be in a larger interval containing the APOL genes rather than be confined to MYH9. Because the risk allele(s) are likely to be common in people with African ancestry, we reasoned that such alleles would be present in the data from the African individuals whose DNA was sequenced in the 1000 Genomes Project (www.1000genomes.org). We therefore used this newly available sequence data to identify polymorphisms within this expanded risk interval that showed large frequency differences between Africans and Europeans in order to test for association with renal disease.
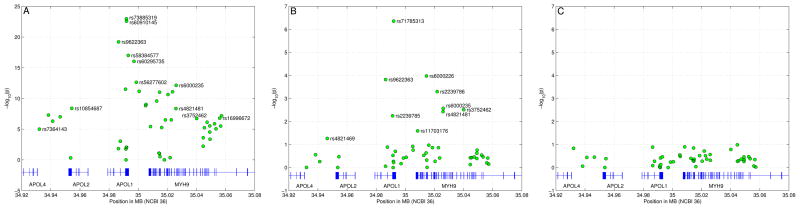
We performed an initial association analysis comparing 205 African-Americans with biopsy proven FSGS but no family history of FSGS with 180 African-American controls. The strongest genetic associations with FSGS were clustered in a 10kb region in the last exon of APOL1, the gene encoding apolipoprotein L-1 (Table S1, Fig. 1A)(14). The strongest signal was obtained for a two-allele haplotype termed “G1” consisting of the two derived non-synonymous coding variants rs73885319 (S342G) and rs60910145 (I384M) in the last exon of APOL1. These two alleles were in perfect LD (r2=1). The G1 haplotype (342G:384M) has a frequency of 52% in FSGS cases and 18% in controls (Table 1, p=1.07×10−23).
Figure 1.
Association analysis in FSGS cohorts with logistic regression for alleles G1 and G2. (A) Results of association beween 205 idiopathic biopsy-proven African American FSGS cases and 180 African American controls using Fisher’s exact test. On the x-axis and y-axis, genomic position and −log10 of the p-values are shown. Also highlighted are SNPs rs4821481 and rs3752462 whose combined risk alleles define the E-1 haplotype(5). (B) SNP associations after conditioning on allele G1 using logistic regression. (C) SNP associations after conditioning on alleles G1 and G2 using logistic regression.
Table 1.
Number and frequencies of APOL1 genotypes and alleles in FSGS and hypertension-attributed ESKD cases and controls.
| Genotype | BWH FSGS Cases | NIH FSGS Cases | Total Cases | Controls | H-ESKD Cases | Controls |
|---|---|---|---|---|---|---|
| WT+WT | 3 | 26 | 29 | 77 | 239 | 409 |
| WT+G1 | 6 | 21 | 27 | 41 | 173 | 250 |
| WT+G2 | 0 | 9 | 9 | 36 | 124 | 155 |
| G1+G1 | 25 | 35 | 60 | 9 | 219 | 41 |
| G1+G2 | 15 | 38 | 53 | 8 | 203 | 50 |
| G2+G2 | 3 | 11 | 14 | 5 | 44 | 18 |
| Total | 52 | 140 | 192 | 176 | 1002 | 923 |
| Allele | ||||||
| G1 Freq. | 0.68 | 0.47 | 0.52 | 0.18 | 0.41 | 0.21 |
| G2 Freq. | 0.19 | 0.25 | 0.23 | 0.15 | 0.21 | 0.13 |
When we performed logistic regression to control for G1, we identified a second strong signal (Table S1, Fig. 1B; p=4.38×10−7). This second signal is a 6 base pair deletion (rs71785313, termed “G2”) close to G1 in APOL1. This deletion removes amino acids N388 and Y389. Due to the proximity of rs73885319, rs60910145, and rs71785313, the two alleles G1 and G2 are mutually exclusive, as recombination between them is very unlikely. Allele G2 has a frequency of 23% in FSGS cases and 15% in controls (Table 1).
After performing regressions controlling for both G1 and G2, we observed no other significant associations (Table S1, Fig. 1C). Conversely, controlling for multiple sets of variants in MYH9 failed to eliminate the APOL1 signal. The LD patterns in this region show that G1 and G2 are in strong LD with variants in MYH9 (Figs S1 and S2). In particular, the MYH9 E-1 haplotype, the best predictor of renal disease in previous studies, is present in most haplotypes containing the G1 or G2 allele. Specifically, E-1 is present in 89% of haplotypes carrying G1 and in 76% of haplotypes carrying G2, explaining the association of MYH9 E-1 with renal disease.
Haplotype frequencies for FSGS cases and controls are shown in Table 1. No difference in FSGS risk was seen when comparing subjects with no risk allele to subjects with one risk allele (G1 or G2, p=0.81, OR=1.04, CI 0.63–2.13). Comparing subjects with zero or one risk allele to subjects with two risk alleles provided an odds ratio for FSGS of 10.5 (CI 6.0–18.4). This analysis supports a completely recessive pattern of inheritance.
Next, we tested association of APOL1 variants and renal disease in a larger cohort of 1030 African-American cases with hypertension-attributed ESKD and 1025 geographically matched African-American controls from the southeastern United States(7). In this cohort, we tested 36 variants chosen based on the strongest signals of positive selection in a broader genomic region. We also tested nearby coding variants including G1 and G2 and putative MYH9 risk single-nucleotide polymorphisms (SNPs). The strongest association was again with rs73885319 (G1 tag SNP; p=1.1 × 10−39) (Table S2). When we controlled for rs73885319 by logistic regression, rs71785313 (G2) again emerged as the strongest association signal (p=8.8×10−18) (Table S2). The statistical significance of the combined signal (p=10−63) was 35 orders of magnitude stronger than for MYH9 SNPs. When we controlled for both G1 and G2, no residual association remained after correction for multiple SNP testing (Table S2). Frequencies for these alleles are shown in Table 1.
With this larger population, we were able to examine the mode of inheritance of renal disease caused by G1 and G2 with greater precision. We partitioned cases and controls by genotype. One risk allele was associated with only a small increase in renal disease risk (odds ratio 1.26, CI 1.01–1.56). Two risk alleles versus zero risk alleles yielded an odds ratio of 7.3 (CI 5.6–9.5). Two risk alleles compared to one risk allele showed an odds ratio of 5.8 (CI 4.5–7.5). Overall, a recessive model best explains these findings and is in agreement with our analysis of the FSGS cohort.
We compared the frequency of G1 and G2 in several HapMap populations using 1000 Genomes sequence data. G1 was present in approximately 40% of Yoruba (from Nigeria in West Africa) chromosomes, but not in any chromosomes from European, Japanese, or Chinese individuals. Similarly, G2 was detected in sequence data from 3 Yoruba subjects, but not in the other three ancestral groups. This distribution data raised the possibility that these variants were selected for in Africa, but not outside of Africa. The high frequency of the disease-associated variants in Yoruba and African-Americans suggests that these variants may confer selective advantage in Africa.
Given the strong evidence for selection previously described in this chromosomal region(9–13), we genotyped G1 and G2 in 180 Yoruba samples from HapMap3 to test these variants for their potential contribution to selection (Table S3). The allele frequencies in Yoruba are 38% for G1 and 8% for G2. We focused on statistical tests that detect selection by evaluating differential degrees of LD surrounding a putatively selected allele compared to the LD around the alternate allele at the same locus(13, 15).
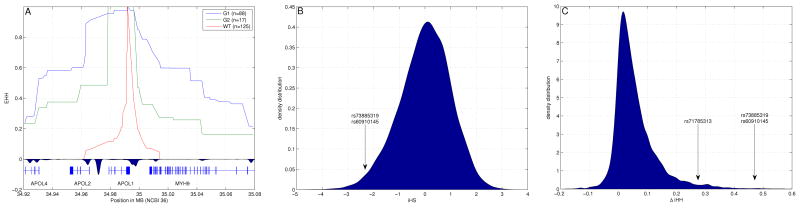
A recent (<10,000 years) selective sweep by a positively selected allele that rises quickly in frequency creates longer patterns of LD around the locus under selection(16). To determine if this is the case for G1 and G2, we computed the Extended Haplotype Homozygosity (EHH)(15) for each one of the two risk alleles and the non-risk allele(Figs. 2A, S3). We also computed the integrated haplotype score (iHS)(13) and ΔiHH(11). The iHS statistic is suited to detect selective sweeps where the selected allele has reached intermediate frequency. |iHS| greater than 2 indicates unusual LD at a locus relative to the rest of the genome, a typical signature of natural selection. As iHS is designed to have a standard normal distribution, its value is significant for the two G1 SNPs (rs73885319 and rs60910145, iHS=−2.45; Fig. 2B). ΔiHH is similar to iHS but tests absolute rather than relative differences in the length of haplotypes(11). ΔiHH was high for G1 SNPs (ΔiHH=0.471 cM, more than 5 s.d. from the mean) and for rs71785313 (G2) (ΔiHH=0.275 cM, 2.6 s.d. from the mean) compared with the genome as a whole, again showing that haplotypes carrying the derived alleles are positively selected (Fig. 2C). Results of multiple tests for selection and population differentiation for the entire region from 34,900–35,100kb (NCBI 36) are reported in Table S4. These same tests in Europeans showed no deviation from neutrality at APOL1.
Figure 2.
Natural selection analysis for the Yoruba population. (A) Extended haplotype homozygosity (EHH) values for the three APOL1 alleles (G1, G2, and WT). We computed EHH after combining Hapmap3 genotype data with our genotype data for alleles G1 and G2 (Supplementary Table 3), shown together with recombination hotspots around APOL1. Although APOL1 and MYH9 are in close physical proximity (20kb apart), they are separated by a recombination hotspot, giving a genetic distance (0.2cM), equivalent to a physical distance of approximately 200kb. (B) Distribution of iHS values in Yoruba, highlighting the iHS value for the SNPs defining G1 (iHS=−2.45). The iHS scores are distributed as a standard normal random variable. Therefore values for which |iHS|>2 are considered suggestive of selection. (C) Distribution of ΔiHH values highlighting the SNPs defining allele G1 and allele G2 (for G1, ΔiHH=0.471 cM, and for G2, ΔiHH=0.275 cM). Values are measured in centimorgans (cM). Raw data are available in Table S4.
Taken together, these data are consistent with the hypothesis that G1 rose quickly to high frequency due to positive selection in Africa. There is less power to show an effect for G2 because of its lower frequency in Yoruba (8%) and the more robust effect of G1 within the same interval, but haplotypes containing G2 show higher degrees of homozygosity than haplotypes that contain neither G1 nor G2, again suggesting positive selection for G2 in Africans (Fig. 2A).
While statistical tests for selection are valuable for identifying haplotypes under selection, only functional tests can convincingly demonstrate the causal allele. Our selection tests indicated that G1 and G2 are on haplotypes that have been strongly selected for in Africa but not in Europe or Asia. G1 and G2 are plausible candidates for the causal variants because they alter the sequence of the encoded protein.
ApoL1 is the trypanolytic factor of human serum that confers resistance to the Trypanosoma brucei brucei (T.b. brucei) parasite(17, 18). T.b. brucei has evolved into two additional subspecies, Trypanosoma brucei rhodesiense and Trypanosoma brucei gambiense, which have both acquired the ability to infect humans(19, 20). T.b. rhodesiense is predominantly found in Eastern and Southeastern Africa, while T.b. gambiense is typically found in Western Africa, though some overlap exists(21). Since these parasites exist only in sub-Saharan Africa, we hypothesized that the APOL1 gene may have undergone natural selective pressure to counteract these trypanosoma adaptations. As an initial test of this hypothesis, we performed in vitro assays in which the trypanolytic potential of the disease-associated ApoL1 proteins was compared with that of the typical ApoL1 protein.
T.b. rhodesiense can infect humans because of a serum resistance-associated protein (SRA) that interacts with the C-terminal helix of ApoL1 and inhibits its anti-trypanosomal activity(18, 22). A recent study showed that mutations and deletions engineered into this helix prevent SRA from binding to ApoL1(23). Intriguingly, one of the G1 sequence variants (I384M) and the 6bp deletion (G2) are located exactly at the SRA binding site in the ApoL1 C-terminal helix.
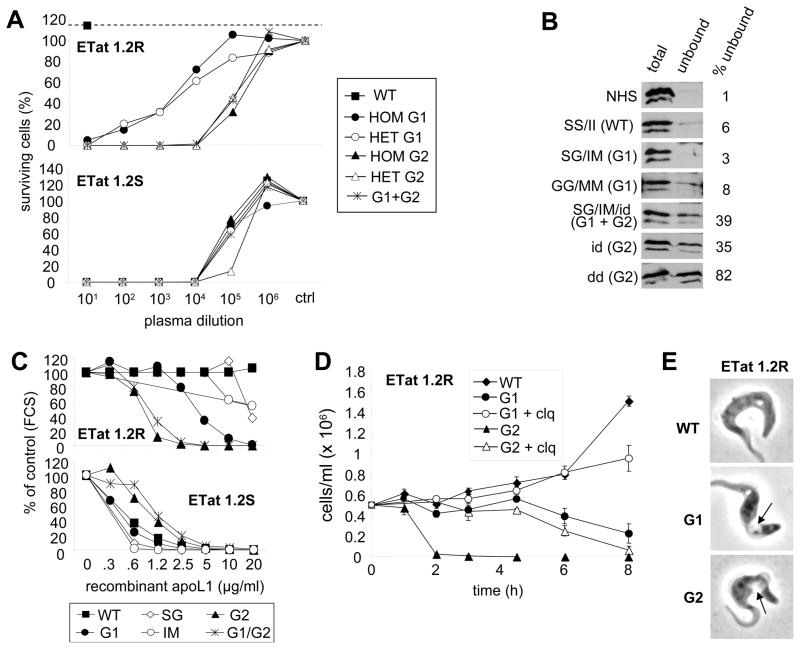
We conducted an analysis of the in vitro lytic potential of 75 human plasma samples with different combinations of G1 and G2 genotypes on T.b. brucei, T.b. rhodesiense, and T.b. gambiense. All 75 plasma samples efficiently lysed T.b. brucei, but none of them lysed T.b. gambiense. Of the 75 samples, 46 lysed SRA-positive T.b. rhodesiense clones, which are typically resistant to lysis by human serum, and all 46 originated from individuals harboring at least one G1 or G2 allele (Table S5). As measured by titration using serial dilution, the lytic potency of these plasmas against SRA-positive T.b. rhodesiense was higher for G2 than for G1, whereas it was similar for both genotypes against SRA-negative parasites (Fig. 3A). While lysis of T.b. rhodesiense by G2 could be explained by the inability of SRA to bind to this mutant, this conclusion did not hold for G1 ApoL1 variants, which SRA could still efficiently bind (Fig. 3B).
Figure 3.
G1 and G2 alleles of ApoL1 kill T.b. rhodesiense. Trypanolytic potential of ApoL1 variants on normal human serum-resistant (SRA+) and normal human serum-sensitive (SRA−) T. b. rhodesiense ETat 1.2 clones. ETat 1.2R is resistant to normal human serum, and ETat 1.2S is sensitive to normal human serum. (A) Titration of trypanolytic activity in human plasma samples after overnight incubation, expressed as % survival compared to fetal calf serum control. (hom, het=homozygous and heterozygous mutations, respectively). (B) ApoL1 content of various plasma samples before and after affinity chromatography through SRA column (NHS=normal human serum; WT=wild type ApoL1; S=serine 342; G=glycine 342; I=isoleucine 384; M=methionine 384; i=insertion of N388/Y389; d=deletion of N388/Y389). (C) Trypanolytic activity of various recombinant ApoL1 variants after overnight incubation, expressed as % survival compared to fetal calf serum (FCS) control. (D) Kinetics of trypanolysis by 20 μg/ml recombinant ApoL1 variants, in the presence or absence of 25 μM chloroquine (clq). (E) Phenotype of ETat1.2R trypanosomes incubated with various recombinant ApoL1 (20 μg/ml; 1h30 and 6h incubation, for G1 and G2 respectively; the arrows point to the swelling lysosome).
We confirmed these results with recombinant ApoL1 proteins. The S342G/I384M (G1) and delN388/Y389 (G2) variants lysed both SRA-negative and SRA-positive T.b. rhodesiense parasites (Fig. 3C), but not T.b. gambiense. While G2 was more potent than G1 against SRA-positive T.b. rhodesiense, the reverse was true on SRA-negative parasites. Recombinant ApoL1 variants with either S342G alone or I384M alone were less lytic against T.b. rhodesiense than when present together, while recombinant ApoL1 engineered to have both G1 and G2 mutations was not more active than mutants with G2 alone (Fig. 3C). As shown in Fig. 3D–E, all measured features of the T.b. rhodesiense lytic process (kinetics, transient inhibition by chloroquine, typical swelling of the lysosome) were similar to those observed on T.b. brucei with either normal human serum or recombinant ApoL1(17). Therefore, deletion of N388/Y389 was necessary and sufficient to prevent interaction with SRA and to confer on ApoL1 the ability to lyse T.b. rhodesiense in vitro, whereas the combination of S342G and I384M was required for maximal ability to lyse T.b. rhodesiense despite remaining bound by SRA. None of the variant forms of ApoL1 lysed T.b. gambiense.
In summary, we have shown that sequence variation in APOL1 contributes to the increased risk of renal disease in African-Americans. Two lines of evidence support this conclusion: (i) the non-synonymous variants coded by G1 and the coding region deletion G2 in APOL1 are the sequence variants showing the strongest association with FSGS and H-ESKD; and (ii) association of renal disease with the MYH9 sequence variants disappears after controlling for the APOL1 risk variants. An important question to be addressed in future studies is how sequence variation in ApoL1 mechanistically contributes to the pathogenesis of kidney disease. The recessive model that best fits our genetic data suggests that ApoL1 is performing a critical role in the kidney that is impaired in the setting of the ApoL1 variants, though toxicity of the ApoL1 variants remains a possibility.
We have shown that both ApoL1 variants lyse a deadly subspecies of Trypanosoma that is normally completely resistant to ApoL1 lytic activity. The G2 mutation prevents the SRA virulence factor produced by T.b. rhodesiense from binding to and inactivating ApoL1. Even 10,000-fold dilutions of plasma containing these mutations (particularly G2) are active against the parasite. This raises the possibility that transfusion of small volumes of plasma, ApoL1-containing HDL particles, or recombinant protein might be effective treatment for trypanosomiasis caused by T.b. rhodesiense.
The kidney-disease associated variants are located on haplotypes that show statistical evidence of natural selection. The lytic activity of the variant proteins against Trypanosoma provides a plausible—albeit still speculative—biological explanation for natural selection. The results are consistent with a heterozygous advantage model since the protective effect against T.b. rhodesiense is dominant whereas the association with renal disease is recessive. Sickle cell disease is a well-established precedent for a model in which mutations conferring heterozygote advantage against a parasitic infection can confer a strong biological disadvantage for homozygotes (24). When present in heterozygous from, certain hemoglobin mutations confer protection against malaria but when homozygous cause severe diseases of the red blood cell (sickle cell disease, thalassemia).
It will be interesting to determine the distribution of these mutations throughout sub-Saharan Africa. In present-day Africa, T.b. rhodesiense is found in the Eastern part of the continent, while we noted high frequency of the trypanolytic variants and the signal of positive selection in a West African population. Changes in trypanosome biology and distribution and/or human migration may explain this discrepancy, or resistance to T.b. rhodesiense could have favored the spreading of T.b. gambiense in West Africa. Alternatively, ApoL1 variants may provide immunity to a broader array of pathogens beyond just T.b. rhodesiense, as a recent report linking ApoL1 with anti-Leishmania activity may suggest(25). Thus, resistance to T.b. rhodesiense may not be the only factor causing these variants to be selected.
The APOL1 risk alleles for renal disease occur in more than 30% of African-American chromosomes. Given their high frequency and their strong effect on disease risk, unraveling the molecular mechanisms by which they contribute to renal injury will be of great importance in understanding and potentially preventing renal disease in individuals of recent African ancestry.
Supplementary Material
Acknowledgments
We thank members of the Broad Institute Medical and Population Genetics Program and review of the manuscript. We thank the study subjects for their participation and Jim Robinson for the help provided accessing the 1000 Genomes project data. We thank X.C. Zhou (NCI) and E. Binns-Roemer (NCI), P. Poelvoorde (ULB), P. Tebabi (ULB) and A. Pays (ULB) for excellent technical assistance. We thank the investigators in the BWH and NIH FSGS genetic studies: William Briggs, Richard Dart, Stephen Korbet, Michelle Mokrzycki, Paul Kimmel, Tejinder Ahuja, Jeff Berns, Eric Simon, Michael Smith, Howard Trachtman, Miriam Michel, Jeffrey R Schelling, Gerald Appel, and Avi Katz. The work at the University of Brussels was supported by the “CIBLES” programme of the Walloon Region, the Fonds National de la Recherche Scientifique and the Interuniversity Attraction Poles programme of the Belgian Science Policy. This work was also supported by grants from the NIH (RO1 DK54931 to M.R.P, K08-DK076868 to D.J.F., RO1 DK066358 and RO1 DK053591 to D.W.B., RO1 HL56266, RO1 DK 070941, and RO1 DK 084149 to B.I.F.), the NIDDK Intramural research program (ZO1 DK043308, to J.B.K.), and the Intramural Research Program of National Cancer Institute, Center for Cancer Research (Project ZIA BC 010022; C.A.W., G.W.N.), the NephCure Foundation (to M.R.P). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U. S. Government. The publisher or recipient acknowledges right of the U. S. Government to retain a non-exclusive, royalty-free license in and to any copyright covering the article. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. All subjects gave informed consent for genetic studies and research was conducted under protocols approved by the appropriate institutional review boards. The NCI, NIH has a patent application pending for chromosome 22 polymorphisms and haplotypes for genetic screening and clinical applications.
References
- 1.Byrne C, Nedelman J, Luke RG. Race, socioeconomic status, and the development of end-stage renal disease. Am J Kidney Dis. 1994 Jan;23:16. doi: 10.1016/s0272-6386(12)80806-7. [DOI] [PubMed] [Google Scholar]
- 2.Satko SG, Freedman BI, Moossavi S. Genetic factors in end-stage renal disease. Kidney Int Suppl. 2005 Apr;S46 doi: 10.1111/j.1523-1755.2005.09411.x. [DOI] [PubMed] [Google Scholar]
- 3.U.S. Renal Data System, National Institutes of Health. National Institute of Diabetes and Digestive and Kidney Diseases. Bethesda, MD: 2009. [Google Scholar]
- 4.Kao WH, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008 Oct;40:1185. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopp JB, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008 Oct;40:1175. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freedman BI, et al. Non-muscle myosin heavy chain 9 gene MYH9 associations in African Americans with clinically diagnosed type 2 diabetes mellitus-associated ESRD. Nephrol Dial Transplant. 2009 Nov;24:3366. doi: 10.1093/ndt/gfp316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman BI, et al. Polymorphisms in the non-muscle myosin heavy chain 9 gene (MYH9) are strongly associated with end-stage renal disease historically attributed to hypertension in African Americans. Kidney Int. 2009 Apr;75:736. doi: 10.1038/ki.2008.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson GW, et al. Dense mapping of MYH9 localizes the strongest kidney disease associations to the region of introns 13 to 15. Hum Mol Genet. 2010 May 1;19:1805. doi: 10.1093/hmg/ddq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008 Mar;40:340. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 10.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007 Oct 18;449:851. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grossman SR, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010 Feb 12;327:883. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 12.Kimura R, Fujimoto A, Tokunaga K, Ohashi J. A practical genome scan for population-specific strong selective sweeps that have reached fixation. PLoS One. 2007;2:e286. doi: 10.1371/journal.pone.0000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006 Mar;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.See supplementary materials in Science online.
- 15.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002 Oct 24;419:832. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 16.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006 Jun 16;312:1614. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Morga D, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005 Jul 15;309:469. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 18.Vanhamme L, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003 Mar 6;422:83. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 19.Gibson W. Will the real Trypanosoma brucei rhodesiense please step forward? Trends Parasitol. 2002 Nov;18:486. doi: 10.1016/s1471-4922(02)02390-5. [DOI] [PubMed] [Google Scholar]
- 20.Gibson WC. Will the real Trypanosoma b. gambiense please stand up. Parasitol Today. 1986 Sep;2:255. doi: 10.1016/0169-4758(86)90011-6. [DOI] [PubMed] [Google Scholar]
- 21.Simarro PP, Jannin J, Cattand P. Eliminating human African trypanosomiasis: where do we stand and what comes next? PLoS Med. 2008 Feb;5:e55. doi: 10.1371/journal.pmed.0050055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xong HV, et al. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998 Dec 11;95:839. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 23.Lecordier L, et al. C-terminal mutants of apolipoprotein L-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS Pathog. 2009 Dec;5:e1000685. doi: 10.1371/journal.ppat.1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams TN. Human red blood cell polymorphisms and malaria. Curr Opin Microbiol. 2006 Aug;9:388. doi: 10.1016/j.mib.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Samanovic M, Molina-Portela MP, Chessler AD, Burleigh BA, Raper J. Trypanosome lytic factor, an antimicrobial high-density lipoprotein, ameliorates Leishmania infection. PLoS Pathog. 2009 Jan;5:e1000276. doi: 10.1371/journal.ppat.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.