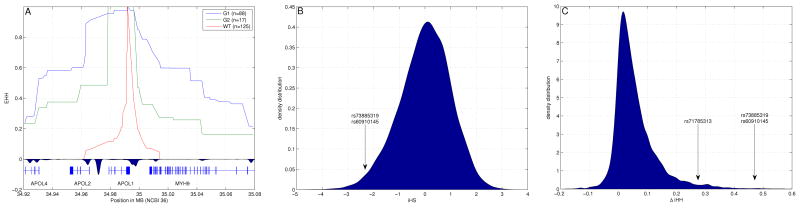
Figure 2.
Natural selection analysis for the Yoruba population. (A) Extended haplotype homozygosity (EHH) values for the three APOL1 alleles (G1, G2, and WT). We computed EHH after combining Hapmap3 genotype data with our genotype data for alleles G1 and G2 (Supplementary Table 3), shown together with recombination hotspots around APOL1. Although APOL1 and MYH9 are in close physical proximity (20kb apart), they are separated by a recombination hotspot, giving a genetic distance (0.2cM), equivalent to a physical distance of approximately 200kb. (B) Distribution of iHS values in Yoruba, highlighting the iHS value for the SNPs defining G1 (iHS=−2.45). The iHS scores are distributed as a standard normal random variable. Therefore values for which |iHS|>2 are considered suggestive of selection. (C) Distribution of ΔiHH values highlighting the SNPs defining allele G1 and allele G2 (for G1, ΔiHH=0.471 cM, and for G2, ΔiHH=0.275 cM). Values are measured in centimorgans (cM). Raw data are available in Table S4.