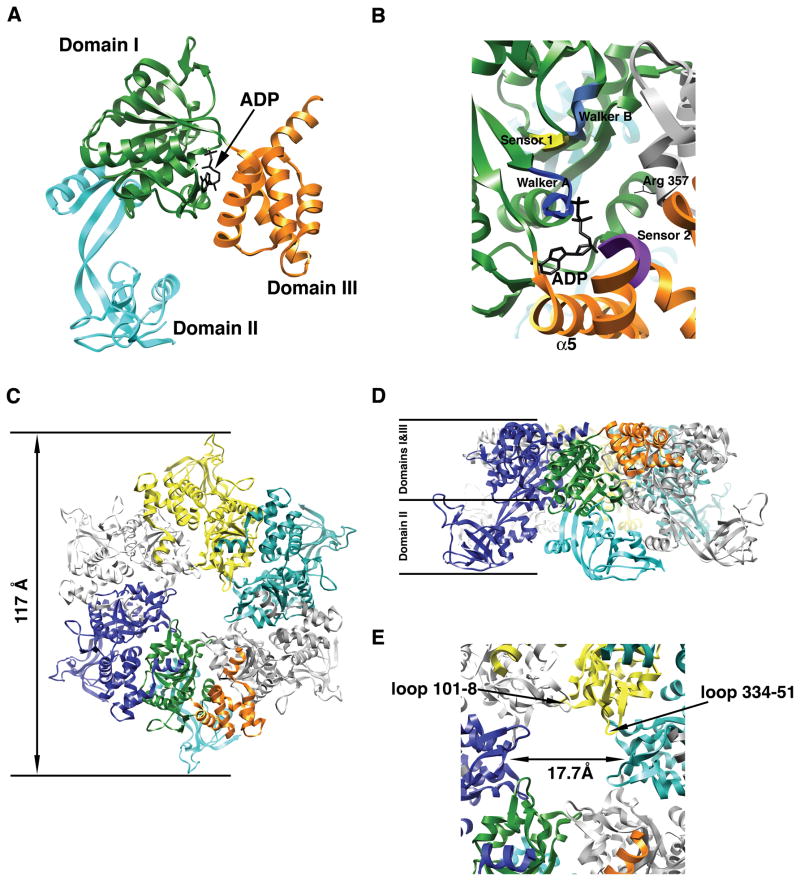
Figure 1. X-ray structure of the human Rvb1 protein.
(A) Ribbon representation of the monomer of human Rvb1 in complex with an ADP molecule. Each domain is represented in a different color. (B) Closed up view of the nucleotide-binding site in the human Rvb1 structure. The elements of the AAA+ module important for nucleotide binding and hydrolysis are represented in different colors and labeled. Domains within the monomer are color coded as in Fig. 1A and the neighboring subunit contributing the “arginine finger” (Arg-357) is colored in grey. (C) Top-view of the ribbon representation of the hexameric ring formed by the human Rvb1 protein in the crystal structure. Each monomer is represented in a different color except the monomer at the bottom that follows the same color code as Fig. 1A for the domains. (D) Side-view of the ribbon representation of the human Rvb1 hexamer. The location of domains I, II and III from the Rvb1 monomers in the hexamer is indicated. Each monomer is represented in a different color and the monomer at the front has its domains colored as in Fig. 1A. (E) Close-up view of the central pore in the human Rvb1 hexamer. The diameter of the pore is indicated and also the two loops within one monomer that are predicted to mediate the binding of ssDNA.