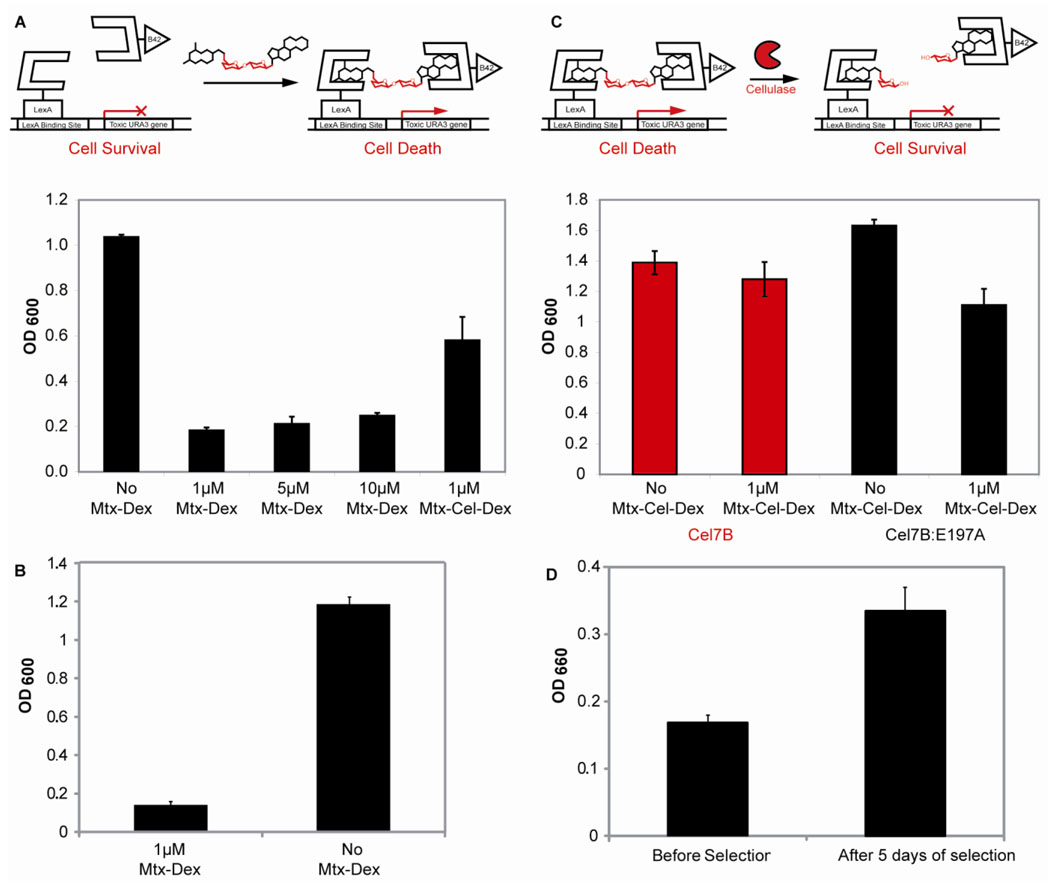
Figure 3.
The chemical complementation URA3 counter selection strain. A. The URA3 reverse yeast three-hybrid system. Absence of small molecule leads to cell survival and an increase in absorption at 600nm. Presence of either 1, 5, 10 µM Mtx-Dex, or 1 µM Mtx-Cel-Dex leads to toxic URA3 reporter gene transcription, cell death, and a decrease of absorption at 600nm. The experiments were carried out in triplicate. Shown is the mean growth and the error bars represent the standard deviation from the mean. B. Reversion rate of the chemical complementation URA3 counter selection strain. Ninety three unique chemical complementation URA3 counter selection conditions (1μ M MTX-Dex) and under non-counter selection conditions (No MTX-Dex) as a control. Shown is the mean growth and the error bars represent the standard error form the mean (p-value<0.0001). C. The chemical complementation URA3 counter selection strain detects cellulase activity. Presence of 1 µM Mtx-Cel-Dex and active cellulase Cel7B leads to cell survival and an increase in absorption at 600 nm. Presence of 1 µM Mtx-Cel-Dex and inactive cellulose Cel7B:E197A leads to cell death and a decrease in absorption at 600nm. As a control, the URA3 counter selection strain was also grown in the absence of Mtx-Cel-Dex and either the presence of Cel7B or Cel7B:E197A. All experiments were carried out in triplicate. Shown is the mean growth and the error bars represent the standard deviation from the mean. D. The chemical complementation URA3 counter selection enriches for cellulase activity after five days of selection. Twenty-two cellulase variants isolated before selection and twenty-two variants isolated after five days of selection were tested for cellulase activity using carboxymethylcellulose (CMC) in crude cell extracts. Increase in aldehyde formation was detected as an increase in absorption at 660nm. Shown is the mean cellulase activity and the error bars represent the standard error form the mean (p-value<0.005).