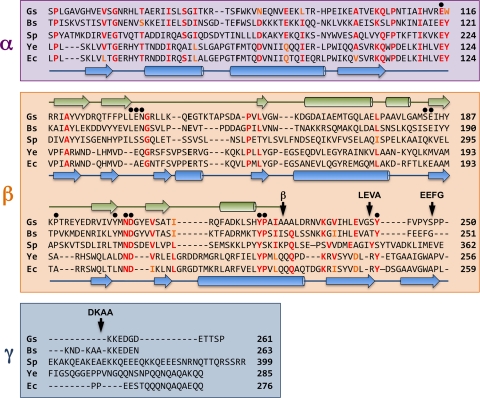
FIG. 3.
Structure-based alignment of the amino acid sequences of the extracytoplasmic regions of DivIB from G. stearothermophilus (Gs), B. subtilis (Bs), and S. pneumoniae (Sp) and the amino acid sequences of FtsQ from Y. enterocolitica (Ys) and E. coli (Ec). The α and β domains and γ tail are highlighted in violet, orange, and blue, respectively; see text for a discussion of the exact location of the β/γ boundary. The arrow labeled β indicates the boundary of the DivIB β domain as defined previously (38) while other arrows labeled LEVA, EEFG, and DKAA indicate the boundaries of the extended β domains used for artificial septal targeting. Black dots highlight residues that were mutated in the current study. The secondary structure of G. stearothermophilus DivIB as determined from solution NMR studies (31) is shown in green above the sequences, while the secondary structure of E. coli FtsQ as determined using X-ray crystallography (36) is shown in blue below the sequences. Residue numbers are indicated at the end of each sequence.