Abstract
The general stress regulon of Bacillus subtilis is controlled by σB, a transcription factor that is activated by physical or nutritional stress. In B. subtilis, each of these two stresses is communicated to the primary σB regulators by distinct pathways. Physical stress activation of σB involves a large-molecular-mass (>106-Da) structure (stressosome) formed by one or more homologous proteins (RsbRA, -B, -C, and -D) onto which the pathway's principal regulators are bound. The RsbR proteins are thought to be potential receptors for stress signaling. Listeria monocytogenes encodes orthologs of σB and its principal regulators; however, unlike B. subtilis, L. monocytogenes appears to use the stressosome pathway for both physical and nutritional stress activation of σB. In the current work, a B. subtilis strain that expressed L. monocytogenes rsbR (rsbRLm) in lieu of B. subtilis rsbR (rsbRBs) was created and was found to display the Listeria phenotype of σB activation following exposure to either physical or nutritional stress. B. subtilis expressing either the RsbR paralog rsbRC or rsbRD, but not rsbRA or rsbRB, as the sole source of RsbR also allowed σB induction following nutritional stress. It is unclear whether the nutritional stress induction seen in these strains is the result of a direct effect of nutritional stress on stressosome activity or a consequence of the background levels of σB activation in these strains and the effects of diminished ATP on the downstream phosphorylation reaction needed to reinactivate σB.
σB is an alternative sigma factor of Bacillus subtilis that directs RNA polymerase to the promoters for the more than 150 genes that make up the bacterium's general stress regulon (GSR) (23, 24, 32, 33, 40). The GSR is activated when exposure to physical (e.g., ethanol or heat or osmotic shock) or nutritional (e.g., glucose or phosphate starvation or O2 limitation) stress initiates a series of reactions which frees σB from an inhibitory association with the anti-σB protein (RsbW) (6, 7, 8, 9). Release of σB from RsbW requires the binding of an additional protein (RsbV) to RsbW (16, 17). In unstressed B. subtilis, RsbV is not able to trigger σB release, due to an RsbW-dependent phosphorylation (17). Phosphorylated RsbV (RsbV-P) is dephosphorylated and reactivated by either of two stress-responsive phosphatases (RsbP or RsbU) (25, 39, 41, 42, 44).
The RsbP phosphatase and an additional protein (RsbQ) are required for nutritional stress activation of σB in wild-type B. subtilis (10, 39). The inducer of RsbPQ phosphatase activity is unknown. Recently, red light has also been shown to be a potential RsbP-dependent activator of σB; however, the details of this activation and its relationship to nutritional stress activation remain to be resolved (5). RsbU, the phosphatase that reacts to physical stress, also requires an additional protein (RsbT) for activity (44). In unstressed B. subtilis, RsbT is sequestered with its primary negative regulator (RsbS) in a large (1.8-MDa) complex formed by a family of homologous proteins: RsbR (RsbRA), YkoB (RsbRB), YojH (RsbRC), and YqhA (RsbRD) (1, 2, 14, 15, 28, 30). This complex of RsbR, -S, and -T proteins has been termed the “stressosome” (28, 30). Physical stress is believed to trigger an RsbT-dependent phosphorylation of both RsbR and RsbS, which allows the release of RsbT and its activation of RsbU (14, 20, 22, 44). The system is reset by RsbX, an additional phosphatase that can dephosphorylate RsbR-P and RsbS-P, allowing their reactivation and the potential resequestration of RsbT (13, 15, 44).
rsbRA is cotranscribed in an 8-gene operon with the σB structural gene and other key σB regulators, while the rsbR paralogs are expressed from diverse sites along the B. subtilis chromosome (2, 43). A fifth RsbR-like paralog has been described (YtvA); it can cofractionate with stressosomes but is unique in that it lacks the RsbT-dependent phosphorylation sites found on the other RsbR paralogs (4, 19). Instead, YtvA carries a light, oxygen, or voltage (LOV) domain. YtvA has been found to enhance σB activation in the presence of blue light, although the mechanism involved is unknown (4, 19, 38).
In vitro, the phosphorylation of RsbR promotes the phosphorylation of RsbS (13, 14, 20). This observation suggests that the stress activation process in vivo could proceed through RsbR to RsbS. As such, the RsbR proteins themselves might serve as targets for intracellular signals that might promote their susceptibility to phosphorylation by RsbT (18, 27). Cryo-electron microscopy (cryo-EM) analyses of stressosomes revealed a structure formed of multiple RsbR molecules with their C-terminal regions arranged as a base onto which RsbS and RsbT are bound (30). Studies of stressosome assembly and composition indicate that individual stressosomes are likely to be mosaics of multiple RsbR paralogs; however, the explicit functions of the multiple RsbR proteins are unclear (15, 28, 35). The N-terminal portions of these proteins, which appear to project outward from the stressosome, are much less conserved than their C-terminal regions (2, 30). This heterogeneity raised the possibility that each of the RsbR paralogs could serve as a receptor for a unique stress signal; however, a study examining the effects of the loss of one or more of the paralogs failed to demonstrate selective responsiveness. The loss of one or several of the RsbR-encoding genes did not prevent σB activation by any of the usual assortment of environmental inducers (2). Only when all of the rsbR genes were deleted did σB activity become unresponsive to physical stress (2). The persistent ability of σB to be induced by diverse physical stresses in the absence of one or more of the RsbR paralogs was interpreted as evidence that the responsiveness of the RsbR proteins to physical stress is overlapping. Given the heterogeneity of the amino termini of the RsbR proteins, their common responsiveness to the same physical stresses could be a consequence of each RsbR paralog responding to one of multiple signals generated by exposure to a given stress. In this view, each of the RsbR proteins would respond to a novel activator, although not in an obvious stress-specific manner.
Listeria monocytogenes encodes a σB ortholog, as well as counterparts of the regulatory proteins that control B. subtilis σB activation in response to physical stress (11, 12, 23, 31). L. monocytogenes σB is activated by both physical and nutritional stress; however, the essential components of B. subtilis σB nutritional stress activation (RsbPQ) are lacking in L. monocytogenes (11, 12). In contrast to B. subtilis, L. monocytogenes is thought to use the components of the physical stress pathway to activate σB following nutrient deprivation (11, 12). To ask whether this unique responsiveness rests with the L. monocytogenes RsbR protein (RsbRLm), we constructed a B. subtilis strain in which the rsbRLm gene was placed within the sigB operon in lieu of the B. subtilis ortholog as the strain's sole source of RsbR. The resulting strain allowed σB activation following either physical or nutritional stress. This induction of σB following nutritional stress, but not physical stress, was blocked if the B. subtilis RsbR (RsbRBs) paralogs were also present. Testing of individual RsbR paralogs for similar properties revealed that RsbRC and RsbRD also permitted σB induction under conditions that normally activated the nutritional stress pathway. As with RsbRLm, σB was not induced by nutritional stress if other RsbR proteins (RsbRA, RsbRB) were present.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All of the B. subtilis strains are derivatives of PY22 (6). The strains and their relevant genotypes are listed in Table 1. Plasmid pARE241 (PA ΔrsbRA1 rsbS rsbT) (35) was used to construct pLAM2, a plasmid formed by placing an L. monocytogenes rsbR gene fragment (amplified from L. monocytogenes LM1061 DNA, a gift of A. Benson, University of Nebraska) into a unique SalI site created immediately downstream of the initiation codon of rsbRA and 3 codons before the rsbRA termination codon. The amplified rsbRLm was cloned “in frame” with the residual B. subtilis rsbRA sequence, adding a Met-Ser-Thr element to its N terminus and a Val-Asp-Leu-Gly-Glu sequence to its C terminus. BSH80 has been described previously (34). BSJ43 (rsbP::spc SPβ ctc::lacZ) is BSA46 (6) transformed with a DNA fragment carrying rsbP disrupted at an internal HindIII site by Spcr (21). BSH163 (ydcE::Cm SPβ ctc::lacZ) is BSH80 transformed with plasmid pARE212 (35). BSH176 (rsbQ::ery SPβ ctc::lacZ) is BSH80 transformed to rsbQ::ery by a DNA fragment carrying rsbQ with an internal NdeI 563-bp segment deleted and replaced by an Eryr cassette (21). BSH192 (rsbQ::TnYLB-1 SPβ ctc::lacZ) is BSH80 with a TnYLB-1 (Kanr) transposon inserted into rsbQ (29). BAR340 (ΔrsbRB2 ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ) and BAR343 [ΔrsbRA1(Cm) ΔrsbRB1::kan ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ] are BAR203 (35) and BAR205 (35), respectively, each transformed to rsbP::spc with DNA from BSJ43. BSH304 (ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ) and BSH305 [ΔrsbRA1(Cm) ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ] are BAR298 (35) and BAR204 (35), respectively, transformed to rsbQ::TnYLB-1 with DNA from BSH192. BSH306 [ΔrsbRA1(Cm) ΔrsbRB1::kan ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ] was constructed by transformation of BSH176 (rsbQ::ery SPβ ctc::lacZ) with DNA from BAR199 [ΔrsbRA1(Cm) ΔrsbRB1::kan ΔrsbRD1::spc SPβ ctc::lacZ] and selection first for Cmr to create BSA217 [ΔrsbRA1(Cm) rsbQ::ery SPβ ctc::lacZ], then for Kanr to form BSH308 [ΔrsbRA1(Cm) ΔrsbRB1::kan rsbQ::ery SPβ ctc::lacZ], and finally for Spcr as the final step in the construction. BSH311 [ΔrsbRA1(Cm) ΔrsbRB2 ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ] was formed in two steps. First, rsbQ::ery was transformed into BAR230 (ΔrsbRB2) from BSH176. The resulting strain, BSA309 (ΔrsbRB2 rsbQ::ery), was then transformed to ΔrsbRA1(Cm) ΔrsbRD1::spc using chromosomal DNA from BSH214 [ΔrsbRA1(Cm) ΔrsbRB1::kan ΔrsbRC1::ery ΔrsbRD1::spc SPβ ctc::lacZ] and selection for Cmr and Spcr. BSA312 [ΔrsbRA1(Cm) ΔrsbRD1::spc rsbQ::ery rsbU::kan SPβ ctc::lacZ] is BSA311 transformed to rsbU::kan with DNA from BSA70 (6). BSH313 [ΔrsbRA1(Cm) ΔrsbRB2 ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ] was made by first transforming BAR230 (ΔrsbRB2) to ΔrsbRC1::ery with DNA from BSH214, followed by transformation to SPβ ctc::lacZ, using Tetr selection. The resulting strain, BSH310 (ΔrsbRB2 ΔrsbRC1::ery SPβ ctc::lacZ) was then transformed to ΔrsbR1(Cm) rsbP::spc with DNA from BAR343. BSH314 [ΔrsbRA1(Cm) ΔrsbRB2 ΔrsbRC1::ery rsbP::spc rsbU::kan SPβ ctc::lacZ] is BSH313 transformed to rsbU::kan with DNA from BSA70 (5). BSH315 [ΔrsbRA1(Cm) ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ] is BSA217 [ΔrsbRA1(Cm) rsbQ::ery SPβ ctc::lacZ] transformed to ΔrsbRD1::spc with DNA from BSH214. BSH316 (ΔrsbRB2 ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ) is BSH309 (ΔrsbRB2 rsbQ::ery) transformed to ΔrsbRD1::spc SPβ ctc::lacZ with DNA from BSH214. BSH317 (ΔrsbRB2 ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ) and BSH318 (PSPAC rsbT rsbP::spc SPβ ctc::lacZ) are BSH310 (ΔrsbRB2 ΔrsbRC1::ery SPβ ctc::lacZ) and BSA419 (PSPAC rsbT SPβ ctc::lacZ) (37), respectively, transformed to rsbP::spc with DNA from BSJ43. BSL20 (ydcE::Cm SPβ rsbQ::TnYLB-1 ctc::lacZ) and BSL24 [PA rsbRD (ΔrsbRA) ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ] are BSH163 (ydcE::Cm SPβ ctc::lacZ) and BAR308 [PA rsbRD (ΔrsbRA) ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc SPβ ctc::lacZ], respectively, transformed to rsbQ::TnYLB-1 with DNA from BSH192. BSL26 [PA rsbRA::rsbRLm(Cm) ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ] is BSH304 transformed with plasmid pLAM2, replacing B. subtilis rsbRA with rsbR from L. monocytogenes. BSL28 [PA rsbRA::rsbRLm(Cm) rsbQ::TnYLB-1 SPβ ctc::lacZ] is BSH80 transformed with DNA from BSL26, selecting for Cmr (PA rsbRA::rsbRLm) and Kanr (rsbQ::TnYLB-1).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant genotype |
|---|---|
| Plasmids | |
| pARE241 | Apr CmrPA ΔrsbRA1 rsbS rsbT |
| pLAM2 | Apr CmrPA rsbRLm rsbS rsbT |
| B. subtilis strains | |
| PY22 | Wild type |
| BAR340 | ΔrsbRB2 ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ |
| BAR343 | ΔrsbRA1(Cm) ΔrsbRB1::kan ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ |
| BSA70 | SPβ ctc::lacZ (Cm Ery) |
| BSH80 | SPβ ctc::lacZ (Tet Ery) |
| BSH176 | rsbQ::ery SPβ ctc::lacZ |
| BSH192 | rsbQ::TnYLB-1 SPβ ctc::lacZ |
| BSH304 | ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ |
| BSH305 | ΔrsbRA1(Cm) ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ |
| BSH306 | ΔrsbRA1(Cm) ΔrsbRB1::kan ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ |
| BSH308 | ΔrsbRA1(Cm) ΔrsbRB1::kan rsbQ::ery SPβ ctc::lacZ |
| BSH311 | ΔrsbRA1(Cm) ΔrsbRB2 ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ |
| BSH312 | ΔrsbRA1(Cm) ΔrsbRB2 ΔrsbRD1::spc rsbQ::ery rsbU::kan SPβ ctc::lacZ |
| BSH313 | ΔrsbRA1(Cm) ΔrsbRB2 ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ |
| BSH314 | ΔrsbRA1(Cm) ΔrsbRB2 ΔrsbRC1::ery rsbP::spc rsbU::kan SPβ ctc::lacZ |
| BSH315 | ΔrsbRA1(Cm) ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ |
| BSH316 | ΔrsbRB2 ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ |
| BSH317 | ΔrsbRB2 ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ |
| BSH318 | PSPAC rsbT rsbP::spc SPβ ctc::lacZ |
| BSJ43 | rsbP::spc SPβ ctc::lacZ |
| BSL20 | ydcE::Cm rsbQ::TnYLB-1 SPβ ctc::lacZ |
| BSL24 | PA rsbRD (ΔrsbRA) ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ |
| BSL26 | PA rsbRA::rsbRLm(Cm) ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ |
| BSL28 | PA rsbR::rsbRLm(Cm) rsbQ::TnYLB-1 SPβ ctc::lacZ |
Growth conditions.
B. subtilis strains were routinely grown in LB (36) with vigorous shaking (250 rpm) at 37°C. Physical stress was initiated by the addition of ethanol or NaCl to a final concentration of 4% or 2.5%, respectively. Nutritional stress was brought on by blocking the electron transport with the addition of sodium azide to 2 mM, by O2 limitation through reducing the rate of culture shaking to 50 rpm, or by allowing the cultures to enter stationary phase in LB. Glucose or phosphate limitation was induced by growth in a synthetic medium (42) with reduced glucose (0.05%) or KH2PO4 (0.18 mM). To elevate the background RsbU activity in the absence of stress, BSH318, carrying rsbT under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter (PSPAC), was grown and repeatedly diluted for 5 generations in LB with IPTG (0.025, 0.05, or 0.1 mM) before samples were taken for analysis of σB activity.
General methods.
β-Galactosidase activities were determined using the chloroform-permeabilized technique of Kenney and Moran (26). Bacillus transformations were performed as described by Yasbin et al. (45).
RESULTS AND DISCUSSION
Expression of rsbRLm in B. subtilis.
L. monocytogenes encodes a σB ortholog, as well as counterparts of the regulatory proteins that control B. subtilis σB activation in response to physical stress, in an operon that is structurally identical to the B. subtilis sigB operon (11, 12, 23, 31). As in B. subtilis, L. monocytogenes σB is activated by both physical and nutritional stress; however, the essential components of Bacillus σB nutritional stress activation (RsbPQ) are lacking in L. monocytogenes (11, 12). In contrast to B. subtilis, L. monocytogenes is thought to use the components of the physical stress pathway to activate σB following both physical stress and nutrient deprivation (11, 12). The RsbR protein encoded within the Listeria sigB operon is highly homologous to B. subtilis RsbRA in its C-terminal half (77% identical amino acids); however, at its N-terminal region, the RsbR segment proposed to be the protein's stress receptor element is much less conserved (22% identical amino acids) (Fig. 1). To ask whether Listeria's responsiveness to nutritional stress might rest with the RsbRLm protein, we constructed a B. subtilis strain in which the rsbRLm gene was placed within the sigB operon in lieu of the B. subtilis ortholog. The remainder of the sigB operon was left intact. The strain also carried disruptions in the paralogous B. subtilis rsbR genes, as well as RsbQ (rsbQ::TnYLB-1), a protein needed for the activity of the B. subtilis nutritional stress pathway's phosphatase. This left only the physical stress pathway intact, with the Listeria RsbR protein as its sole RsbR element.
FIG. 1.
Sequence comparison of RsbR of L. monocytogenes and RsbRA of B. subtilis. The sequence of the L. monocytogenes RsbR protein is given below that of the B. subtilis ortholog. The numbering is that of the B. subtilis protein. Identical amino acids are shaded. The conserved threonine sites for phosphorylation (T171 and T205) are indicated by arrows.
Given the high degree of homology of the C-terminal region of RsbRLm with the corresponding regions of the B. subtilis RsbR proteins, it seemed likely that RsbRLm would be able to form stressosomes in B. subtilis that could incorporate the B. subtilis regulators. To verify this, a preliminary experiment was undertaken in which crude extracts from the rsbRLm-expressing strain were examined by velocity centrifugation. As was done previously in similar experiments conducted on strains expressing other single B. subtilis rsbR genes (35), stressosome formation was estimated by the movement of RsbS into fast-sedimenting complexes. As anticipated, the rsbRLm-expressing strain displayed the presence of B. subtilis RsbS in high-molecular-weight fractions, indicative of stressosome formation (data not shown).
The rsbRLm-expressing strain and a congenic strain expressing B. subtilis rsbRA were next grown in LB and exposed to 4% ethanol or 2.5% NaCl, conditions which normally activate the B. subtilis stressosome-dependent pathway for σB induction (42). As illustrated in Fig. 2, σB-dependent reporter gene activity in both strains was initially low but rapidly increased following exposure to either stress. This reveals that the RsbRLm protein is able to function in B. subtilis, interacting with the B. subtilis σB regulators to restrict σB activity during growth and allow its activation when the bacterium is exposed to physical stress. Apparently, the stress signals to which the RsbRLm protein responds are also generated by stress in B. subtilis.
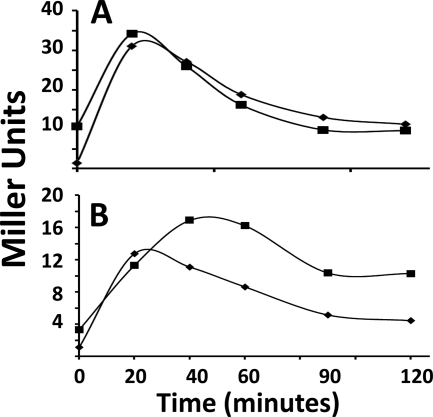
FIG. 2.
σB induction by ethanol or salt in B. subtilis expressing either rsbRLm or rsbRALm. B. subtilis strains BSH304 (ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ) (RsbRA+ strain) (♦) and BSL26 [PA rsbRA::rsbRLm(Cm) ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ] (RsbRLm strain) (▪) were grown in LB and subjected to 4% ethanol (A) or 2.5% NaCl (B) at the zero time point. Samples were taken at the indicated intervals and assayed for σB-dependent β-galactosidase activity.
The rsbRLm and rsbRALm strains were then either exposed to sodium azide or allowed to enter stationary phase, conditions which normally require the nutritional stress pathway for σB activation in wild-type B. subtilis. The rsbRLm-expressing strain, but not the strain expressing the Bacillus ortholog, displayed significant σB induction under these conditions (Fig. 3). The ability of stressosomes formed from Listeria RsbR, when present as the cell's sole RsbR protein, to allow σB activation under nutrient-limiting conditions invites the question of whether σB activation under these circumstances might be influenced by the presence of the B. subtilis paralogs, proteins that would presumably form composite stressosomes with Listeria RsbR. To examine this, the Listeria rsbR gene was substituted for B. subtilis rsbRA in a strain that still encoded the three RsbR paralogs but lacked the nutritional stress pathway phosphatase. When this strain was subjected to nutritional stress by azide treatment or entry into stationary phase, the previous RsbRLm-dependent induction of σB failed to occur (Fig. 3). Thus, the presence of the other RsbR proteins can block the σB induction observed in the strain that expressed RsbRLm alone.
FIG. 3.
σB induction following nutritional stress in B. subtilis expressing either rsbRLm or rsbRABs. B. subtilis strains BSH304 (ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ) (RsbRA+ strain) (♦), BSL26 [PA rsbRA::rsbRLm(Cm) ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ] (RsbRLm strain) (▪), and BSL28 [PA rsbRA::rsbRLm(Cm) rsbQ::TnYLB-1 SPβ ctc::lacZ] (▵) were treated with azide (2 mM) at time zero (A) or grown to stationary phase (arrow) in LB (B). Samples were taken at the indicated intervals and assayed for σB-dependent β-galactosidase activity.
RsbRC and RsbRD allow σB activation following nutritional stress.
The observation that Listeria RsbR allows σB activation following nutritional stress when it is the sole RsbR protein present in B. subtilis raises the question of whether this is a unique characteristic of Listeria RsbR or whether other RsbR proteins share this property. Nutrient stress activation of σB via the physical stress pathway is not evident in B. subtilis expressing the full complement of RsbR proteins; however, as was seen with RsbRLm, the ability of a single RsbR protein to allow σB activation could be blocked in the presence of the other RsbR proteins within the cell.
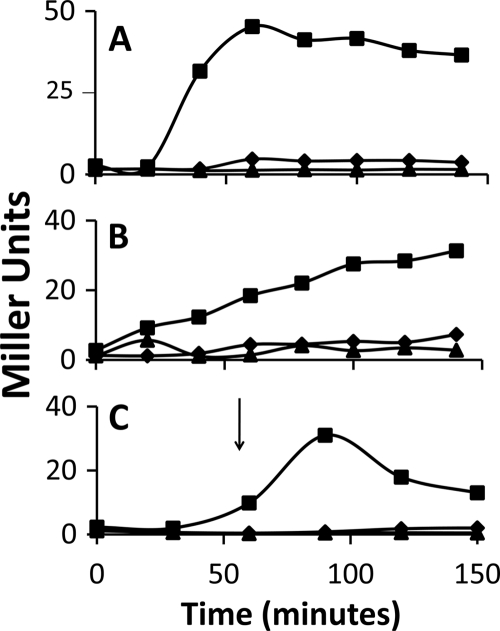
In order to test the possibility that individual RsbR paralogs might be capable of allowing σB activation during nutritional stress, strains which carried a σB-dependent reporter gene (ctc::lacZ strains), lacked the nutritional stress phosphatase (i.e., rsbP::spc or rsbQ::ery strains), and carried disruptions within all but one of the rsbR genes were constructed. The rsbR gene that remained in each strain was expressed from its normal locus. These strains, as well as an RsbPQ− strain expressing all of the rsbR genes, were grown in LB and exposed to ethanol to verify the presence of an intact physical stress pathway in each. All of the strains, as expected, showed induction of σB in response to ethanol (Fig. 4A). When these same strains were allowed to enter stationary phase, those expressing either rsbRC or rsbRD alone also displayed σB activation (Fig. 4B). Neither the parental strain nor the rsbRA- or rsbRB-expressing strains responded to entry into stationary phase with the induction of σB. The presence of both RsbRA and RsbRB is not necessary to prevent σB activation under presumed nutritional stress. The stationary-phase induction in the rsbRC-expressing strains was blocked if either rsbRA or rsbRB alone was present (Fig. 5). Similar results were seen in the rsbRD-expressing strain (data not shown). Curiously, a strain which expresses both rsbRC and rsbRD, although still responsive to nutritional stress, was only half as effective in activating σB as a strain expressing each of these genes singly (Fig. 5). Apparently, the presence of both RsbRC and RsbRD forms a more effective barrier to σB activation following nutritional stress. Perhaps composite stressosomes, even when composed of RsbR paralogs that can singly permit σB activation under this condition, are less adept at allowing the response when these proteins are both present.
FIG. 4.
σB induction by ethanol or entry into stationary phase in B. subtilis strains with single RsbR proteins. B. subtilis strains BSJ43 (rsbP::spc SPβ ctc::lacZ) (○), BSH304 (ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ) (+), BSH305 [ΔrsbRA1(Cm) ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ] (▵), BSH306 [ΔrsbRA1(Cm) ΔrsbRB1::kan ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ] (▪), and BAR343 [ΔrsbRA1(Cm) ΔrsbRB1::kan ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ] (♦) were grown in LB and either subjected to 4% ethanol (A) or allowed to enter stationary phase (B). Samples were taken at the indicated intervals and assayed for σB-dependent β-galactosidase activity. Ethanol was added at the zero time point in panel A. The entry into stationary phase is indicated by the arrow in panel B.
FIG. 5.
RsbU-dependent σB activation in stationary phase by B. subtilis with novel RsbR combinations. B. subtilis strains BSH308 [ΔrsbRA1(Cm) ΔrsbRB1::kan rsbQ::ery SPβ ctc::lacZ] (▴), BSH311 [ΔrsbRA1(Cm) ΔrsbRB2 ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ] (♦), BAR343 [ΔrsbRA1(Cm) ΔrsbRB1::kan ΔrsbRC1::ery rsbP::spc SPβ ctc::lacZ] (▪), BSH315 [ΔrsbRA1(Cm) ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ] (✖), and BSH316 (ΔrsbRB2 ΔrsbRD1::spc rsbQ::ery SPβ ctc::lacZ) (×) were grown to stationary phase (indicated by the arrow) in LB. Samples were taken at the indicated intervals and assayed for σB-dependent β-galactosidase activity.
In order to verify that the stationary-phase induction of σB in the rsbRC- and rsbRD-expressing strains is dependent on the physical stress pathway, a disruption of this pathway's phosphatase (rsbU::kan) was introduced into the rsbRC and rsbRD strains, creating rsbRC- or rsbRD-expressing variants that lacked both the RsbU and RsbPQ phosphatases. When these strains were allowed to enter stationary phase in LB, σB-dependent reporter gene activity was no longer induced (data not shown).
RsbRLm, RsbRCLm, and RsbRDLm respond to multiple stimuli associated with nutritional stress activation.
The ability of B. subtilis expressing rsbRLm, rsbRCLm, or rsbRDLm as the sole rsbR gene to activate σB upon entry into stationary phase may indicate that the strains are responding to nutritional stress or are being affected by an unappreciated stress signal that is generated at the end of exponential growth and not nutritional stress per se. The nutritional stress pathway in B. subtilis, defined by its dependence on the RsbPQ phosphatase, is activated coincident with exposure to a number of agents or culture conditions that have as their common feature the ability to cause a drop in ATP levels (42, 46). To determine whether the induction of σB activity in the rsbRLm-, rsbRCLm-, and rsbRDLm-expressing strains is similarly affected by such treatments, σB-dependent reporter gene activity was examined following treatment with sodium azide, restrictive O2 conditions, or limitation for glucose or phosphate. The ability of the rsbRLm-, rsbRCLm-, and rsbRDLm-expressing strains to respond to these potential inducers was compared to the response of similarly treated strains that expressed all of the RsbR proteins and lacked either the physical stress (rsbU::kan) or the nutritional stress (rsbP::spc) phosphatase. The results of this exercise are summarized in Table 2. As expected, the RsbP− strain, lacking the nutritional stress pathway's phosphatase but otherwise wild type, displayed very low σB activity under all of the conditions that normally induce the nutritional stress pathway. In contrast, the RsbU− strain, lacking the physical stress phosphatase but with an intact nutritional stress pathway, exhibited σB activity levels that were substantially greater than those seen in the RsbP− strain under these same conditions. The strains lacking the nutritional stress phosphatase but expressing rsbRLm, rsbRCLm, or rsbRDLm as the sole RsbR protein displayed levels of σB activity that were similar to or greater than the levels seen in the strain with an intact nutritional stress pathway.
TABLE 2.
σB-dependent β-galactosidase activity under the indicated conditions
| Condition | β-Galactosidase activity (Miller units)a |
||||
|---|---|---|---|---|---|
| RsbP− strain | RsbU− strain | RsbRLm strain | RsbRC+ strain | RsbRD+ strain | |
| Stationary phase | 0.6 ± 0.2 | 19.4 ± 5.6 | 25.7 ± 5.4 | 21.8 ± 2.0 | 22.1 ± 1.0 |
| Sodium azide treatment | 0.4 ± 0.4 | 4.7 ± 0.8 | 23.6 ± 1.4 | 47.9 ± 14.4 | 32.3 ± 8.9 |
| O2 deprivation | 2.5 ± 0.7 | 7.4 ± 2.9 | 26.4 ± 1.2 | 15.0 ± 2.4 | 27.0 ± 4.2 |
| Glucose limitation | 1.7 ± 0.5 | 43.3 ± 7.2 | 22.9 ± 1.3 | 29.0 ± 5.3 | 30.4 ± 4.9 |
| Phosphate limitation | 3.8 ± 1.3 | 45.8 ± 10.1 | 21.8 ± 1.2 | 48.1 ± 15.9 | 47.6 ± 7.4 |
Values are averages ± standard deviations from three separate determinations. RsbP− strain, BSJ43; RsbU− strain, BSA70; RsbRLm strain, BSL26; RsbRC+ strain, BSH306 (stationary phase, sodium azide treatment, and O2 deprivation) or BSH311 (glucose/phosphate limitation); RsbRD+ strain, BAR343 (stationary phase, sodium azide treatment, and O2 deprivation) or BSH313 (glucose/phosphate limitation). Stationary-phase values were obtained at 30 min after exponential growth. Sodium azide treatment and O2 deprivation readings were obtained 40 min after initiating the treatments. Glucose and phosphate limitation values were determined 60 min after growth slowed in glucose or phosphate limiting medium.
Although these results are consistent with the notion that all three of these RsbR proteins (RsbRLm, RsbRCLm, and RsbRDLm) can allow σB induction following stresses that are similar to those that activate the σB nutritional stress pathway, a previous study revealed that rsbRD is expressed at a relatively low level compared to that of the other rsbR genes (35). This raises the formal possibility that if entry into stationary phase or other treatments which inhibit growth give rise to a physiological state in which protein turnover is accelerated, the strains that express solely rsbRD might be more susceptible than the other rsbR-expressing strains to becoming functionally RsbR− and no longer able to form the stressosomes needed to hold RsbT inactive. To address the possible complication raised by the low expression level of rsbRD, we repeated the σB induction analysis using a strain in which the rsbRD sequence had been recombined into the B. subtilis chromosome, in lieu of rsbRA, at the sigB operon. This created a strain that is similar to the strain that expressed rsbRLm from this site, allowing rsbRD expression under the control of the rsbRA regulatory elements as the cells' sole source of RsbR. In the previous study mentioned above (35), expression of rsbRD from this site increased RsbD-dependent stressosomes 3-fold, to a level that is over 60% of that seen when RsbRA-dependent stressosomes are formed. This is a level of stressosome formation equivalent to that seen when RsbRB is the sole RsbR source (35). Figure 6 depicts the σB-dependent reporter gene activity in three RsbPQ− strains: one that expresses all of the rsbR genes and two others that express either rsbRA or rsbRD from within the sigB operon. The strain with rsbRD expressed from sigB, but neither of the other two strains, induced σB activity following azide treatment (Fig. 6A), O2 limitation (Fig. 6B), or entry into stationary phase (Fig. 6C). The level of σB activity seen in the strain with rsbRD at sigB was equal to or greater than that seen in the strain with rsbRD expressed from its normal locus (Fig. 4; Table 2). Taken together, the data argue that all three of these RsbR proteins (RsbRLm, RsbRCLm, and RsbRDLm) can allow σB induction following exposure to any of a number of conditions that are normally associated with nutritional stress activation of σB.
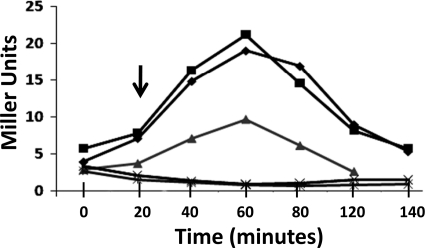
FIG. 6.
RsbRD-dependent nutritional stress activation of σB. B. subtilis strains BSL20 (ydcE::Cm rsbQ::TnYLB-1 SPβ ctc::lacZ) (▴), BSH304 (ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ) (♦), and BSL24 [PA rsbRD (ΔrsbRA) ΔrsbRB2 ΔrsbRC1::ery ΔrsbRD1::spc rsbQ::TnYLB-1 SPβ ctc::lacZ] (▪) were grown in LB and subjected to sodium azide (2 mM) (A) or O2 limitation (B) at the beginning of the experiment (time zero) or allowed to enter stationary phase at the time point indicated by the arrow (C). Samples were taken at the indicated intervals and assayed for σB-dependent β-galactosidase activity.
σB activation during nutritional stress in a strain with elevated background σB activity.
The activation of σB during nutritional stress in B. subtilis expressing rsbRLm, rsbRCLm, or rsbRDLm as its sole source of RsbR may be a direct result of nutritional stress on stressosome activity. However, an alternative explanation of the data, which invokes a more dynamic view of stressosome activity and σB regulation, is possible. It is known that σB activity is very high during growth in B. subtilis strains lacking RsbX, the phosphatase responsible for reactivating RsbR-P and RsbS-P (6). This observation suggests that even in the absence of overt stress there is likely to be a background level of RsbR/RsbS phosphorylation, RsbT release, and activation of σB. Such a circumstance would include the dephosphorylation of RsbV-P, which would then require phosphorylation, at the expense of ATP, to reinactivate RsbV and allow the sequestration of σB into a complex with RsbW (3, 17).
The background level of σB activity in strains expressing single rsbR genes, particularly rsbRLm, is severalfold higher than that seen in the strain expressing all of the B. subtilis rsbR genes or rsbRA alone. Such heterogeneity might be a consequence of the inherent biochemical properties of each of these proteins and/or disparities in the background levels of the signaling molecules to which each responds. Regardless of the basis of these differences in σB activity, it is plausible that higher background levels of σB activity could have consequences when the strains with these higher levels experience nutritional stress. The strains with inherently higher levels of RsbV-P dephosphorylation might be more prone to activate σB when ATP levels fall and the cell's ability to phosphorylate RsbV would presumably be diminished. Early in vitro studies revealed a close correlation among the concentration of ATP required for efficient RsbW-mediated phosphorylation of RsbV, inhibition of RsbV/RsbW complex formation, and σB-directed transcription (3). Prior to the discovery of the RsbPQ phosphatase, the effect of a decline in ATP levels on RsbW kinase activity was, in fact, suggested as the device responsible for activation of σB by nutritional stress (3). In the alternative model, the signal to which Listeria RsbR responds is not generated by both physical and nutritional stress. Instead, RsbRLm might respond solely to signals generated by physical stress but exhibits a sufficiently high “steady-state” level of σB activation to allow σB activity to be more sensitive to changes in ATP levels than that seen in strains with RsbR proteins that have lower background levels of σB activity.
To test the possibility that elevated background activity in the physical stress pathway might allow heightened σB activity during periods of nutritional stress, a B. subtilis strain (BSH318) expressing the activator (RsbT) of the RsbU phosphatase under the control of an IPTG-inducible promoter (PSPAC::rsbT) was grown and allowed to enter stationary phase in the presence of various concentrations of IPTG. Induction of PSPAC::rsbT increases the expression of rsbT relative to that of its primary negative regulator (rsbS), thereby allowing enhanced activity of the physical stress pathway in the absence of stress (37). Figure 7 illustrates that the addition of increasing amounts of IPTG leads to corresponding increases in σB activity during growth, which rises further when the cultures enter stationary phase. Presumably, the stationary-phase elevation in σB activity is due to the decreased availability of ATP for rephosphorylation of the RsbV that had become dephosphorylated by the increased RsbU phosphatase activity. Although entry into stationary phase enhanced σB activity in this B. subtilis strain with heightened background levels of RsbR-P dephosphorylation, the degree of this enhancement, at least under the conditions used in this experiment, was relatively modest (Fig. 7) compared to that seen when strains with stressosomes formed from RsbRLm, RsbRCLm, or RsbRDLm entered stationary phase (Fig. 3, 4, and 6). If the nutritional stress induction of σB activity in the RsbRLm, RsbRCLm, or RsbRDLm strain is a consequence of heightened σB background activity, the robust induction seen in these strains, compared to that seen when σB activity is artificially elevated, suggests that there are additional properties associated with the stressosome-associated process that are lacking when the elevated σB activity is generated by merely raising RsbU phosphatase activity.
FIG. 7.
RsbU-dependent σB activation in stationary phase by B. subtilis strains with induced RsbU activity. B. subtilis BSH318 (PSPAC rsbT rsbP::spc SPβ ctc::lacZ) was grown in LB without IPTG (♦) or supplemented with 0.025 mM (▪), 0.05 mM (▴), or 0.1 mM (×) IPTG to stationary phase (arrow). Samples were taken at the indicated intervals and assayed for σB-dependent β-galactosidase activity.
Regardless of whether nutritional stress allows σB activation by directly targeting the RsbRLm protein or the ability of RsbW to maintain RsbV's phosphorylation state in the presence of RsbRLm, the finding that the RsbRLm protein, but not its B. subtilis counterpart, allows σB activation during nutritional stress offers the Listeria RsbR protein itself as a plausible basis for the observation that σB can be activated by the stressosome-associated pathway following nutritional stress in Listeria but not B. subtilis. Sorting out the target for the nutritional stress activation will ultimately require the development of assays to directly monitor the effects of nutritional stress on the phosphorylation state of the RsbR proteins themselves. If they are responding directly to signals generated by either physical or nutritional stress, their level of phosphorylation would be expected to increase under either of these conditions. However, if the RsbR proteins do not respond to signals generated directly by nutritional stress, their phosphorylation state should remain unchanged under this condition.
The Listeria RsbR protein, although highly homologous at its C terminus to B. subtilis RsbRA and its paralogs, has a novel N-terminal region. Assuming that this region is the target for stress signaling, its uniqueness suggests that it responds to a potentially novel input. The ability of the Listeria RsbR protein to function in B. subtilis, restricting σB activity during growth but allowing its activation following stress, argues both that it can productively interact with the B. subtilis regulators and that the signals to which it responds are generated by stress in B. subtilis as well as in Listeria. This opens the possibility that the signaling molecules that activate the L. monocytogenes stressosome could be sought and studied in the more experimentally tractable B. subtilis, rather than in Listeria itself. RsbRLm, as a novel RsbR variant that can apparently interact with the B. subtilis stressosome components, could also serve as an additional vehicle to study B. subtilis stressosome activity. If, for example, RsbRLm responds directly to a signal generated by nutritional stress, inhibition of its activation of σB by other RsbR proteins would support the notion of stressosomes as composites of multiple RsbR species in which the presence of each component influences the activities of the others (30). The stressosome then becomes a device for integrating multiple stress signals to allow σB activation only when a critical stress threshold is reached (30). Given the diversity of the amino termini of the RsbR paralogs, the specific signaling molecules to which each reacts may be unique and potentially generated by distinct stress-responsive elements within the cell. If this is so, the individual RsbR paralogs could then serve as a gauge of the effects of stress on multiple cell components. In this view, the stressosome may represent not only a device that responds to overall stress levels but also one that integrates regulatory inputs from diverse sources to allow σB activation only when each of several cell components has been affected by stress to provide its input. Uncovering the specific signals to which each of the RsbR proteins responds will be an important next step toward exploring this model.
Footnotes
Published ahead of print on 8 October 2010.
REFERENCES
- 1.Akbar, S., C. M. Kang, T. A. Gaidenko, and C. W. Price. 1997. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24:567-578. [DOI] [PubMed] [Google Scholar]
- 2.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. M. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 4.Avila-Perez, M., K. J. Hellingwerf, and R. Kort. 2006. Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J. Bacteriol. 188:6411-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avila-Perez, M., J. B. van der Steen, R. Kort, and K. J. Hellingwerf. 2010. Red light activates the σB-mediated general stress response of Bacillus subtilis via the energy branch of the upstream signaling cascade. J. Bacteriol. 192:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, A. K., and W. G. Haldenwang. 1992. Characterization of a regulatory network that controls σB expression in Bacillus subtilis. J. Bacteriol. 174:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor σB by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brody, M. S., K. Vijay, and C. W. Price. 2001. Catalytic function of an α/β hydrolase is required for energy stress activation of the σB transcription factor in Bacillus subtilis. J. Bacteriol. 183:6422-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaturongakul, S., and K. J. Boor. 2006. σB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 72:5197-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, C.-C., M. D. Yudkin, and O. Delumeau. 2004. Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis. J. Bacteriol. 186:6830-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, C.-C., R. J. Lewis, R. Harris, M. D. Yudkin, and O. Delumeau. 2003. A supermolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 49:1657-1669. [DOI] [PubMed] [Google Scholar]
- 15.Delumeau, O., C.-C. Chen, J. W. Murray, M. D. Yudkin, and R. J. Lewis. 2006. High-molecular-weight complexes of RsbR and paralogues in the environmental signaling pathway. J. Bacteriol. 188:7885-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delumeau, O., R. J. Lewis, and M. D. Yudkin. 2002. Protein-protein interactions that regulate the energy stress activation of sigma (B) in Bacillus subtilis. J. Bacteriol. 184:5583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eymann, C., D. Becher, J. Bernhardt, K. Gronau, A. Klutzny, and M. Hecker. 2007. Dynamics of protein phosphorlation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics 7:3509-3526. [DOI] [PubMed] [Google Scholar]
- 19.Gaidenko, T. A., T.-J. Kim, A. L. Weigel, M. S. Brody, and C. W. Price. 2006. The blue-light receptor YtvA acts as an environmental stress signaling pathway of Bacillus subtilis. J. Bacteriol. 188:6387-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaidenko, T. A., X. Yang, Y. M. Lee, and C. W. Price. 1999. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the stress signaling pathway of Bacillus subtilis. J. Mol. Biol. 288:29-39. [DOI] [PubMed] [Google Scholar]
- 21.Guérout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 22.Hardwick, S. W., J. Pane-Farre, O. Delumeau, J. Marles-Wright, J. W. Murray, M. Hecker, and R. Lewis. 2007. Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis. J. Biol. Chem. 282:11562-11572. [DOI] [PubMed] [Google Scholar]
- 23.Hecker, M., J. Pane-Farre, and U. Volker. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram positive bacteria. Annu. Rev. Microbiol. 61:215-236. [DOI] [PubMed] [Google Scholar]
- 24.Hecker, M., W. Schumann, and U. Voelker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 25.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenney, T. J., and C. P. Moran, Jr. 1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor of Bacillus subtilis. J. Bacteriol. 169:3329-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, T.-J., T. A. Gaidenko, and C. W. Price. 2004. In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 186:6124-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, T.-J., T. A. Gaidenko, and C. W. Price. 2004. A multi-component protein complex mediates environmental stress signaling in Bacillus subtilis. J. Mol. Biol. 341:135-150. [DOI] [PubMed] [Google Scholar]
- 29.Le Breton, Y. L., and N. P. Mohapatra. 2006. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl. Environ. Microbiol. 72:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marles-Wright, J., T. Grant, O. Delumeau, G. van Duinin, S. J. Firbank, P. J. Lewis, J. W. Murray, J. A. Newman, M. B. Quin, P. R. Race, A. Rohou, W. Tichelaar, M. van Heel, and R. J. Lewis. 2008. Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science 322:92-96. [DOI] [PubMed] [Google Scholar]
- 31.Pane-Farre, J., R. J. Lewis, and J. Stulke. 2005. The RsbRST stress module in bacteria: a signaling system that may interact with different output modules. J. Mol. Biotechnol. 9:65-76. [DOI] [PubMed] [Google Scholar]
- 32.Petersohn, A., J. Bernhardt, U. Gerth, D. Hoper, T. Koburger, U. Voelker, and M. Hecker. 1999. Identification of σB-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181:5718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 34.Reeves, A., U. Gerth, U. Voelker, and W. G. Haldenwang. 2007. ClpP modulates the activity of the Bacillus subtilis stress response transcription factor, σB. J. Bacteriol. 189:6168-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reeves, A., L. Martinez, and W. Haldenwang. 2010. Expression of, and in vivo stressosome formation by, single members of the RsbR protein family in Bacillus subtilis. Microbiology 156:990-998. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Scott, J. M., and W. G. Haldenwang. 1999. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor σB. J. Bacteriol. 181:4653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, N., N. Takaya, T. Hashino, and A. Nakamura. 2007. Enhancement of a σB-dependent stress response in Bacillus subtilis by light via the YtvA photoreceptor. J. Gen. Appl. Microbiol. 53:81-88. [DOI] [PubMed] [Google Scholar]
- 39.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 40.Voelker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Voelker, R. Schmid, H. Mach, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741-752. [DOI] [PubMed] [Google Scholar]
- 41.Voelker, U., A. Voelker, and W. G. Haldenwang. 1996. Reactivation of the Bacillus subtilis anti-σB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J. Bacteriol. 178:5456-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wise, A. A., and C. W. Price. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J. Bacteriol. 177:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 45.Yasbin, R. E., G. A. Wilson, and F. E. Young. 1973. Transformation and transfection of lysogenic strains of Bacillus subtilis 168. J. Bacteriol. 113:540-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, S., and W. G. Haldenwang. 2005. Contributions of ATP, GTP, and redox state to nutritional stress activation of the Bacillus subtilis σB transcription factor. J. Bacteriol. 187:7554-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]