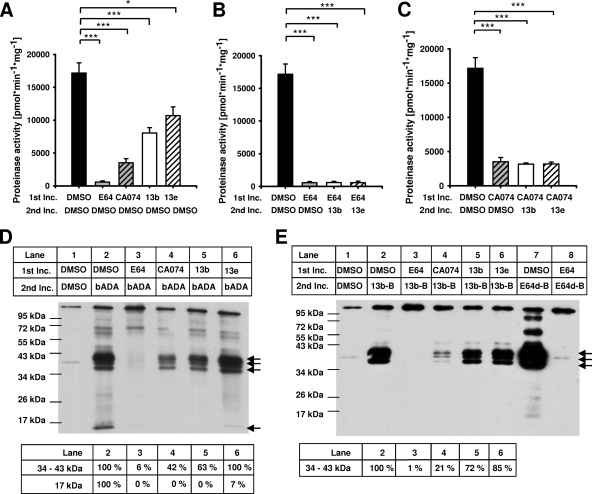
FIG. 2.
Fluorescence proteinase activity assays and active-site labeling experiments with biotin-tagged cysteine cathepsin inhibitors. (A to C) For fluorescence proteinase activity assays, protein lysates that had been obtained from stationary-phase promastigotes were preincubated in a first incubation step (1st Inc.) with DMSO, 100 μM E64, 100 μM CA074, 200 μM compound 13b, or 200 μM compound 13e. In a second incubation step (2nd Inc.), protein lysates were incubated with either DMSO, 200 μM compound 13b, or 200 μM compound 13e. Proteinase activities were determined by proteolytic degradation of the fluoropeptide Z-Phe-Arg-AMC. Values represent means ± SEMs of six independent experiments. (D and E) For active-site labeling, protein lysates were preincubated in a first incubation step (1st Inc.) with DMSO, 100 μM E64, 50 μM CA074, 200 μM compound 13b, or 200 μM compound 13e. In a second incubation step (2nd Inc.), protein lysates were incubated with the biotin-labeled inhibitor bADA (200 μM), 13b-B (200 μM), or E64d-B (2 μM). Controls lacking biotin-tagged inhibitors were incubated with DMSO in both incubation steps. Arrows indicate cysteine cathepsin-specific proteinase bands between 34 and 43 kDa and at 17 kDa. The intensities of these bands were densitometrically determined (tables below the blots). The values for DMSO-preincubated samples labeled with bADA or 13b-B in the second incubation step were set equal to 100%. *, P ≤ 0.05; ***, P ≤ 0.001.