Abstract
The pyrazinamide (PZA) analog 5-chloropyrazinamide (5-Cl PZA) is active against mycobacterial species, including PZA-resistant strains of Mycobacterium tuberculosis. In M. smegmatis, overexpression of the type 1 fatty acid synthase (FAS I) confers resistance to 5-Cl PZA, a potent FAS I inhibitor. Since M. tuberculosis and M. bovis cannot tolerate FAS I overexpression, 5-Cl PZA resistance mutations have yet to be described for tubercle bacilli. In an attempt to identify other factors that govern the activity of 5-Cl PZA, we selected for 5-Cl PZA-resistant isolates from a library of transposon-mutagenized M. smegmatis isolates. Here, we report that increased expression of the M. smegmatis pyrazinamidase PzaA confers resistance to 5-Cl PZA and susceptibility to PZA in M. smegmatis, M. tuberculosis, and M. bovis. In contrast, while ectopic overexpression of the M. tuberculosis pyrazinamidase PncA increases PZA susceptibility, this amidase does not mediate resistance to 5-Cl PZA. We conclude that PncA-independent turnover of 5-Cl PZA represents a potential mechanism of resistance to this compound for M. tuberculosis, which will likely translate into enhanced PZA susceptibility. Thus, countersusceptibility can be manipulated as a resistance-proofing strategy for PZA-based compounds when these agents are used simultaneously.
Pyrazinamide (PZA) is a first-line sterilizing antituberculosis drug which, in combination with isoniazid (INH) and rifampin (RIF), has allowed a reduction in the duration of tuberculosis (TB) chemotherapy from 9 months to 6 months (7). The global surge in multidrug-resistant (MDR) (resistant to INH and RIF) and extensively drug-resistant (XDR) (resistant to INH, RIF, at least 1 quinolone, and at least 1 second-line injectable antituberculosis drug) strains of Mycobacterium tuberculosis are threatening efforts to control TB-associated morbidity and mortality (5, 8). With MDR TB therapy often exceeding 2 years and many cases of XDR TB being incurable with existing treatment regimens, the potential to eradicate this once-manageable disease is rapidly devolving to that of the preantibiotic era. Thus, development of novel antituberculosis drugs is essential to regain control over this global health threat.
PZA is a prodrug of the pharmacologically active agent pyrazinoic acid (PA), which is formed upon deamidation by pyrazinamidase (PZase) (11). Deamidation is essential for cellular accumulation of this compound, as the neutral amide is not readily retained within cells (24). Most organisms, including several mycobacterial species, are intrinsically resistant to PZA due in part to insufficient deamidation (11, 19), inefficient uptake (15), and/or the presence of efficient PA efflux mechanisms (24). The unique susceptibility of M. tuberculosis to PZA has been attributed to the presence of an active PZase and a deficiency in PA efflux (11, 24). Thus far, the only defined mechanism of clinical resistance to PZA is through loss-of-function mutations in the gene encoding the major PZase, pncA (17). In M. smegmatis, PZA deamidation can be catalyzed by PncA and, alternatively, by PzaA (10), an amidase with a broader substrate range (2). Increased conversion of PZA to PA through ectopic expression of either PZase enhances the susceptibility of M. tuberculosis to PZA and also converts the inherently highly PZA-resistant M. smegmatis to PZA susceptible (10).
Spontaneous target site mutations that confer resistance to PA have yet to be described for any mycobacterial species. In the absence of genetic clues, models for the mode of action of this compound have been drawn from studies of the physiologic responses of mycobacteria to PA and its analogs. Importantly, conditions that promote accumulation of PA, such as acidic pH (13, 24) and inhibition of efflux (23), markedly increase its potency, indicating that PA likely has an intracellular target. Based on the pKa and acidic-pH dependence of PA, it has been suggested that this weak acid works via a proton ionophore-type mechanism that directly causes uncoupling of the cellular membrane potential (22, 25). However, it is important to note that PZA and many of its derivatives, such as n-propyl pyrazinoate, PA, and 5-Cl PZA, inhibit mycobacterial type 1 fatty acid synthase (FAS I) in whole-cell and cell-free assays (14, 27). Moreover, synthetic overexpression of mycobacterial FAS I in M. smegmatis confers resistance to 5-Cl PZA (26). Thus, the aforementioned uncoupling effect might be a consequence of a disruption in membrane synthesis due to inhibition of FAS I.
Interestingly, 5-Cl PZA is active against PZA-resistant M. tuberculosis and all other mycobacteria that have been tested, in part because the activity of the compound does not require deamidation (4). In fact, the nonamide form of 5-Cl PZA, 5-Cl pyrazinoic acid (5-Cl PA), is approximately 8-fold less potent against various mycobacterial species (4). Thus, deamidation of 5-Cl PZA to 5-Cl PA represents a potential mechanism for resistance to 5-Cl PZA. However, it is important to note that mycobacterial strains deficient in PZase activity do not show elevated susceptibility to the compound, indicating that wild-type levels of PZase do not appear to modulate 5-Cl PZA susceptibility (4). Furthermore, with the narrow substrate range of PncA (2), it is unclear whether 5-Cl PZA can serve as a substrate for the amidase.
Thus far, mutations in the FAS I locus have not been identified in spontaneously occurring 5-Cl PZA-resistant isolates of M. smegmatis, indicating involvement of additional loci in resistance to the agent (26). Moreover, M. tuberculosis mutant strains with resistance to 5-Cl PZA have not been described. Here, we show that ectopic expression of PzaA in M. smegmatis, M. tuberculosis, and M. bovis confers resistance to 5-Cl PZA and is concomitant with enhanced susceptibility to PZA. In contrast, we find that elevated expression of the M. tuberculosis amidase PncA does not confer resistance to 5-Cl PZA, yet it does enhance PZA susceptibility. Thus, while deamidation represents a potential mechanism for resistance to 5-Cl PZA, such resistance cannot be mediated by native PncA. Further, we find that PZA susceptibility correlates with inhibition of FAS I. Together, these observations demonstrate that 5-Cl PZA and other non-PZase-requiring PZA derivatives show great promise as therapeutic agents for treatment of PZA-resistant TB.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The mycobacterial strains used in this study are described in Tables 1 and 2 and were cultivated with Middlebrook 7H9 and 7H10 growth media (Difco). For M. smegmatis, 0.2% (wt/vol) dextrose and 0.05% (vol/vol) tyloxapol (Sigma-Aldrich) were added to the growth media. For M. tuberculosis and M. bovis, media were supplemented with 0.1 volume of oleic acid-albumin-dextrose-catalase (OADC) (Difco), 0.05% (vol/vol) tyloxapol, and 0.5% (wt/vol) glycerol. Escherichia coli strains HB101 (3), TOP10 (Invitrogen), and DH5αλpir (18) were used for propagation of plasmids, phasmids, and cosmids and were cultivated using LB medium (Difco). When necessary, carbenicillin (50 μg ml−1), kanamycin (40 μg ml−1), and hygromycin (150 μg ml−1) were used. PZA and PA were obtained from Sigma-Aldrich. 5-Cl PZA and 5-Cl PA were synthesized as previously described (4).
TABLE 1.
PZA/5-ClPZA turnover and MICs in M. smegmatis mutants
| M. smegmatis strain | Description | PZA/5-Cl PZA turnover (nmol/min/ml of cells [OD600]) | MIC (μg/ml) |
Reference(s) | |
|---|---|---|---|---|---|
| 5-Cl PZA | PZA | ||||
| mc2155 | Wild-type strain | 0.85 ± 0.1 | 25 | >4,000 | 19, 24 |
| mc22612 | Spontaneous 5-Cl PZAr mutant | 32 ± 3 | 125 | >4,000 | 24 |
| mc27031 | mc2155 MSMEG_1088::magellan4 | 100 ± 15 | 125 | 150 | This work |
| mc27032 | mc2155 ΔMSMEG_1088 | 65 ± 5 | 125 | 150 | This work |
| mc27034 | mc27031 ΔMSMEG_1090 | 1.2 ± 0.1 | 25 | >4,000 | This work |
| mc27035 | mc2155 ΔMSMEG_1090 | 0.95 ± 0.15 | 25 | >4,000 | This work |
| mc27036 | mc22612 ΔMSMEG_1090 | 1.2 ± 0.2 | 25 | >4,000 | This work |
| mc27037 | Spontaneous 5-Cl PZAr mutant | 100 ± 15 | 125 | 150 | This work |
| mc27038 | mc2155 attBL5::PTc::pzaAMsmeg | 34 ± 1 | 125 | >4,000 | This work |
Transposon mutagenesis, mutant selection, and insertion site identification.
A library of 105 independent M. smegmatis::magellan4 mutants was generated as previously described using the mariner-based transposon delivery phage phAE180 (12, 16). Mutant strains were pooled in 7H9 medium, and 4 × 105 CFU were spread on medium containing 100 μg ml−1 5-Cl PZA. The plates were incubated at 37°C for 3 days. The magellan4 insertion sites were identified as previously described (16). In brief, genomic DNA was extracted, digested with BssHII, ligated with T4 DNA ligase, and used for transformation of E. coli DH5αλpir (16) to kanamycin resistance. Purified plasmid DNA was sequenced by the DNA Core Facility of the Albert Einstein College of Medicine using the magellan4 primers m4R1 (5′-TAGACCGAGATAGGGTTGAG-3′) and m4F1 (5′-GCATCGCCTTCTATCGCCTTC-3′). Transposon-chromosome junctions were identified by comparing relevant plasmid-derived sequences with the M. smegmatis mc2155 genome sequence using the NCBI genome BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Spontaneous 5-Cl PZA-resistant mutants were selected by plating 4 × 107 CFU on 7H10 (0.2% dextrose) containing 100 μg ml−1 5-Cl PZA as previously described (26).
Construction of mycobacterial mutant strains.
Temperature-sensitive specialized transducing phasmids (1) were constructed and used for allelic-exchange-mediated deletion of the MSMEG_1088, MSMEG_1090 (pzaA), and MSMEG_6506 (pncA) genes in M. smegmatis. DNA segments of approximately 1 kb flanking the gene of interest were amplified by PCR, digested with Van91I, and ligated to compatible fragments of the counterselectable vector p0004S (a gift from T. Hsu) (unpublished data). Allelic-exchange plasmids were selected and propagated following transformation of E. coli TOP10 to hygromycin resistance. The sequences of the inserted DNA fragments were verified. The allelic-exchange plasmids were digested with PacI and ligated to PacI-digested phAE159 (12). The ligation products were packaged using Gigapack Gold III (Stratagene, La Jolla, CA) and transduced to E. coli HB101 for propagation as cosmids. Cosmid DNA was purified and electroporated into M. smegmatis for phage propagation at the permissive temperature (30°C). Subsequently, M. smegmatis strains were transduced at the nonpermissive temperature (37°C), and recombinant strains with integration of the allelic-exchange marker were selected on LB agar plates containing hygromycin. Integration of the allelic-exchange marker at the locus of interest was verified by PCR using one primer that anneals within the exchange marker and one primer that anneals distal to the genomic region that was involved in allelic exchange.
For amidase expression, strains were transformed by electroporation with the mycobacterial integrative expression vector pTIC6a (9) containing pncA from M. tuberculosis H37Rv or pzaA from M. smegmatis. Transformants were selected on supplemented 7H10 medium containing kanamycin. PncA or PzaA expression was induced by addition of 50 ng ml−1 anhydrotetracycline (6, 9).
Assessment of susceptibility to PZA analogs.
The MICs for PZA and 5-Cl PZA were determined using standard broth susceptibility assays (4). Exponentially growing cells were diluted to 106 CFU ml−1 in supplemented 7H9 medium containing various concentrations of PZA or 5-Cl PZA. Where indicated, culture media were adjusted to pH 5.2 (for M. smegmatis) or pH 6.0 (for M. tuberculosis and M. bovis). Cultures were incubated for 2 days (for M. smegmatis) or 11 days (for M. tuberculosis and M. bovis) at 37°C. The MIC was defined as the lowest concentration of compound required to inhibit visible growth. Resistance frequencies (defined as the ratio of resistant CFU to input CFU) were determined by plating 107 to 108 CFU on solid medium containing the respective compounds. Single-cell suspensions were used for accurate assessment of resistance frequencies and were obtained by passing each culture through a 5-μm filter (Millipore).
Pyrazinamidase assays.
Pyrazinamidase activity was measured using a quantitative discontinuous whole-cell Fe(II)-dependent colorimetric assay modified from that of Wayne (21). Exponentially growing cells were harvested by centrifugation (3,450 × g; 10 min) and resuspended in sterile deionized water containing 0.05% tyloxapol. PZA or 5-Cl PZA was added to 2 mM, and samples were incubated at 37°C with shaking. At various intervals, aliquots were removed and clarified by centrifugation (13,000 × g; 5 min; 4°C). Ferrous ammonium sulfate was added to a final concentration of 20 mM. The samples were incubated at 4°C for 30 min and clarified by centrifugation (as described above), and the optical density at 468 nm (OD468) was determined immediately. PA and 5-Cl PA concentrations were determined based on absorbance at 468 nm with millimolar extinction coefficients of 0.68 and 0.47, respectively. One unit is defined as the amount of cellular material (A600 units) required for the deamidation of 1 nmol of substrate in 1 min in a 1-ml reaction volume. The reaction described above for spectrophotometric assays was also used as a modified simple pyrazinamidase (Wayne) test (21) using the culture medium with PZA and iron as the background color.
Determination of type 1 fatty acid synthase activity.
To test whether PZA susceptibility in M. smegmatis correlates with FAS I inhibition, strain mc27031 was treated with PZA and synthesis of the primary product of FAS I (palmitic acid) was measured. Strains were grown to mid-log phase (OD600, 0.5), cells were harvested by centrifugation (3,450 × g; 10 min; 4°C), resuspended to an OD600 of 0.2 in 10 ml 7H9 medium (pH 5.1) containing 2.5 mg ml−1 PZA, and incubated for 2 h at 37°C. [1-14C]acetate (1 μCi ml−1) was added, and the cultures were incubated for an additional 2 h. Cells were harvested by centrifugation (3,450 × g; 10 min; 4°C). Fatty acid preparation and analysis were performed as previously described (28). In brief, cells were harvested by centrifugation and washed twice with H2O. The cell pellets were extracted with chloroform-methanol (2:1 [vol/vol]) to yield a soluble lipid fraction, which was evaporated under nitrogen before saponification using a 25% methanolic KOH solution for 3 h at reflux. Samples were then acidified to a pH of <2. Fatty acids were extracted with chloroform and evaporated under nitrogen. The residue was derivatized using an Alltech kit (catalog no. 18036) to yield UV-absorbing p-bromophenacyl fatty acids. The p-bromophenacyl fatty acid esters were then analyzed by high-performance liquid chromatography using a reverse-phase C18 column, a diode array detector, and an IN/US β-RAM flowthrough beta-gamma radiation detector as described previously (26, 28). [14C]palmitate was quantified and compared to the level of [14C]palmitate of the untreated sample to yield relative incorporation of [14C]acetate as a measure of FAS I inhibition (27).
RESULTS
Identification of mutations that confer 5-Cl PZA resistance.
Consistent with previous reports (26), spontaneous M. smegmatis mutant strains resistant to 100 μg ml−1 of 5-Cl PZA (4-fold the MIC) could be isolated at a frequency of approximately 10−6. However, the frequency of spontaneous resistance to 250 μg ml−1 5-Cl PZA was beyond the limit of detection (<2.5 × 10−8). Interestingly, when strains resistant to 100 μg ml−1 5-Cl PZA were plated on medium containing 250 μg ml−1 5-Cl PZA, resistant mutants were still not obtained. Similarly, when the wild-type M. smegmatis strain mc2155 was plated on medium containing 750 μg ml−1 PA (5-fold the MIC) or 1 mg ml−1 5-Cl PA (2-fold the MIC), the frequency of spontaneous resistance was beyond the limit of detection (<2.5 × 10−8). Such a low incidence of PA and 5-Cl PA resistance suggests that single target site mutations that confer resistance to these compounds are extremely rare.
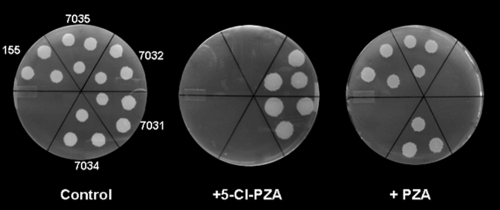
In order to isolate 5-Cl PZA-resistant strains with readily tractable mutations, 4 × 105 CFU from a pooled library of 105 independent M. smegmatis::magellan4 mutants were plated on medium containing 100 μg ml−1 5-Cl PZA. Resistant strains were isolated at a frequency of 10−4, indicating that specific magellan4 insertion mutations can confer resistance to this compound in M. smegmatis. Similar to the previously described spontaneous 5-Cl PZA-resistant mutants (26), the selected M. smegmatis::megellan4 mutants were resistant to 100 μg ml−1, while susceptibilities to 5-Cl PA and PA were unchanged (500 and 375 μg ml−1, respectively). Interestingly, while the wild-type M. smegmatis strain was resistant to >4,000 μg ml−1 PZA (2), the M. smegmatis::megellan4 5-Cl PZA-resistant mutants were found to be susceptible to 150 μg ml−1 PZA (Table 1 and Fig. 1).
FIG. 1.
Counterresistance to 5-Cl PZA and PZA in M. smegmatis. Shown are the 5-Cl PZA resistance and PZA susceptibilities of M. smegmatis strains on solid media following disruption or deletion of MSMEG_1088 (mc27031 and mc27032, respectively) and after deletion of MSMEG_1090 (pzaA) from strains mc27031 (mc27034) and 155 (mc27035).
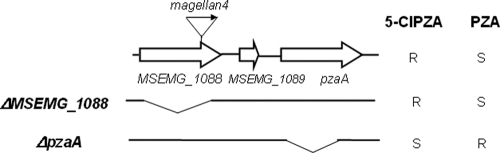
Of 15 5-Cl PZA-resistant mutants that were analyzed, all harbored the transposon insertion at identical positions, approximately 100 bp from the 3′ end of MSMEG_1088 (Fig. 2). MSMEG_1088 encodes an uncharacterized glutamyl-tRNA amidotransferase homolog and is located 243 bp upstream of the pzaA gene (MSMEG_1090) in the same putative operon (Fig. 2). To determine whether the 5-Cl PZA resistance of the M. smegmatis MSMEG_1088::magellan4 strain (mc27031) was due to loss of function of MSMEG_1088, an intact copy of the allele was introduced under the control of a tetracycline-inducible promoter (6, 9). Under inducing conditions, this construct did not restore 5-Cl PZA susceptibility (the MIC was still 125 μg ml−1), suggesting that the 5-Cl PZA resistance of strain mc27031 was not due to loss of MSMEG_1088 function. In contrast, deletion of pzaA (Fig. 2) in this strain (resulting in strain mc27034) restored full susceptibility to 5-Cl PZA (Fig. 1 and Table 1). Finally, transformation of the wild-type strain mc2155 with a plasmid containing an inducible copy of pzaA (strain mc2 7038) (Table 1) resulted in 5-Cl PZA resistance. Thus, 5-Cl PZA resistance can be mediated by increased expression of pzaA.
FIG. 2.
Physical map of the M. smegmatis 5-Cl PZA resistance locus and corresponding resistance (R)/susceptibility (S) phenotypes of locus mutants. The block arrows represent open reading frames, the small arrow represents the magellan4 insertion in MSMEG_1088 in strain mc27031, and the horizontal lines marked ΔMSMEG_1088 and ΔpzaA represent deletions that were introduced in various strains described in the text.
Increased pyrazinamidase activity in 5-Cl PZA-resistant mutants.
As the mycobacterial MIC for 5-Cl PA is much higher than that for 5-Cl PZA (4) and elevated expression of PZase leads to enhanced PZA susceptibility (2), we measured the PZase activity of M. smegmatis mc27031 and an M. smegmatis ΔMSMEG_1088 mutant strain (mc27032) (Fig. 2). Both mutants were found to convert PZA to PA at a higher rate than the wild-type strain (mc2155) (Table 1), suggesting that these mutations confer increased expression of the pzaA gene product. Interestingly, PZase activity was greater in mc27031 than in mc27032, but this increased level of amidase activity did not translate into a further increase in the MIC for 5-Cl PZA (Table 1).
Consistent with a role for PZase as a mediator of 5-Cl PZA resistance, spontaneous 5-Cl PZA-resistant mutant strains of M. smegmatis were also found to have increased levels of this activity (Table 1). In these strains, the level of PZase activity correlated with the degree of susceptibility to PZA. While a newly isolated spontaneous 5-Cl PZA-resistant mutant strain (mc27037) had a high level of PZase and was susceptible to 150 μg ml−1 PZA (Table 1), strain mc27038 (encoding an inducible copy of pzaA) and strain mc22612 (a previously described 5-Cl PZA-resistant mutant strain) (26) showed lower levels of PZase and were not susceptible to PZA (Table 1).
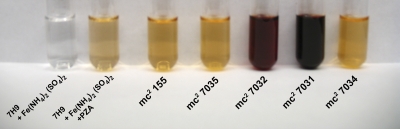
The differences in amidase activity conferred by MSMEG_1090 (Table 1) could also be detected in a modified simple pyrazinamidase (Wayne) test (21) showing color change to dark-cherry discoloration for the 5-Cl PZA-resistant strains (Fig. 3).
FIG. 3.
Simple pyrazinamidase (Wayne) assay in broth. Two 5-Cl PZA-resistant M. smegmatis strains and derivatives with MSMEG_1090 (pzaA) deleted (Table 1) were tested for pyrazinamidase activity as described in Materials and Methods.
PZA inhibits the activity of FAS I in PZA-susceptible M. smegmatis strains.
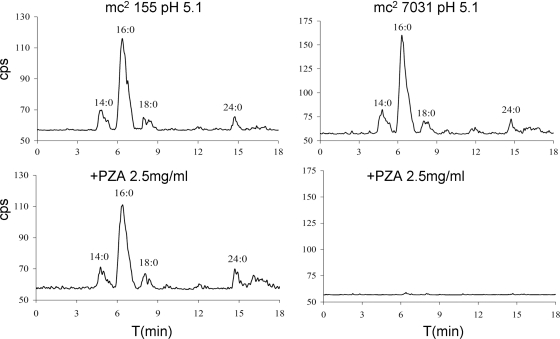
We previously found that PZA susceptibility correlates with FAS I inhibition (26, 27). To assess this concept for M. smegmatis, strain mc27031 was treated with PZA, and synthesis of the primary product of FAS I (palmitic acid) was measured. As shown in Fig. 4, PZA treatment inhibited the FAS I activity of strain mc27031 yet had no effect on this activity in the wild-type strain, mc2155. Further, 1,500 μg ml−1 5-Cl PZA was required to inhibit FAS I in strain mc27031 compared to 250 μg ml−1 for FAS I inhibition in mc2 155. These results demonstrate a correlation between FAS I inhibition and susceptibility to PZA and 5-Cl PZA in M. smegmatis.
FIG. 4.
High-performance liquid chromatography (HPLC) analysis of C16:0 to C26:0 fatty acids from PZA-treated M. smegmatis strains mc2155 and mc27031. As described in Materials and Methods, the strains were cultivated in 7H9 medium containing 0.2% dextrose, 0.05% tyloxapol, pH 5.1; treated with 2.5 mg/ml PZA for 2 h; and then pulsed with [1-14C]acetate for an additional 2 h. Cells were collected by centrifugation, and lipids were extracted, saponified, reextracted, derivatized to p-bromophenacyl esters, and analyzed for 14C content by reverse-phase HPLC.
Ectopic expression of PzaA and PncA affects PZA and 5-Cl PZA deamidation and susceptibility in tubercle bacilli.
Consistent with a previous report (2), ectopic expression of either PncA or PzaA increased the PZA susceptibility of M. bovis and M. tuberculosis during incubation under acidic conditions (Table 2). Interestingly, while tubercle bacilli are naturally tolerant of high concentrations of PZA at neutral pH, increased expression of PncA and PzaA also conferred PZA susceptibility during incubation under neutral pH conditions (Table 2).
TABLE 2.
Effect of PncA and PzaA expression on PZA and 5-Cl PZA turnover and MIC in tuberculous bacilli
| Strain | Characteristica | 5-Cl PZA |
PZA |
Reference | ||||
|---|---|---|---|---|---|---|---|---|
| Turnover (nmol/min/ml of cells [OD600]) | MIC μg/ml |
Turnover (nmol/min/ml of cells [OD600]) | MIC μg/ml |
|||||
| pH 6 | pH 6.8 | pH 6 | pH 6.8 | |||||
| M. tuberculosis H37Ra | Attenuated mutant of H37Rv | 0.15 ± 0.01 | 25 | 25 | 0.23 ± 0.01 | >1,000 | 50 | |
| mc27092 | H37Ra attBL5::PTc::pzaAMsmeg | 27 ± 1 | 200 | 100 | 20 ± 1 | 62.5 | 25 | This work |
| mc27093 | H37Ra attBL5::PTc::pncAMtb | 0.096 ± 0.002 | 25 | 25 | 2.9 ± 1 | 62.5 | 50 | This work |
| M. bovis BCG-Pasteur | Attenuated mutant of M. bovis | 0.11 ± 0.01 | 12.5 | 12.5 | 0.015 ± 0.001 | >1,000 | >1,000 | |
| mc27091 | BCG-Pasteur attBL5::PTc::pzaAMsmeg | 24 ± 1 | 200 | 50 | 16 ± 1 | 12.5 | 62.5 | This work |
| mc27099 | BCG-Pasteur attBL5::PTc::pncAMtb | 0.080 ± 0.01 | 12.5 | 12.5 | 3.5 ± 0.1 | 12.5 | 62.5 | This work |
Msmeg, M. smegmatis; Mtb, M. tuberculosis.
As was observed for M. smegmatis, ectopic expression of PzaA in tubercle bacilli also mediated resistance to 5-Cl PZA, which correlated with the turnover of this compound (Table 2). In contrast, increased expression of PncA had no measurable effect on 5-Cl PZA susceptibility and turnover (Table 2), revealing that PncA does not recognize 5-Cl PZA as a substrate.
To assess whether PZA and 5-Cl PZA have synergistic or antagonistic effects on each other, the MICs for these compounds were determined during coadministration to strains resistant to each. For M. bovis BCG (naturally resistant to PZA), the MIC for 5-Cl PZA was maintained at 12.5 μg/ml regardless of the presence of 100 μg/ml PZA. Similarly, for M. tuberculosis mc27092 (H37Ra attBL5::PTc::pzaAMsmeg; resistant to 5-Cl PZA), the MIC for PZA was 25 μg/ml regardless of the presence of 25 μg/ml 5-Cl PZA. Thus, coadministration of PZA and 5-Cl PZA is neutral and does not affect the susceptibility of tubercle bacilli to the active analog of the pair. Despite the observation that M. bovis has a loss of function mutation in the gene encoding PncA and is resistant to high levels of PZA, we find that this bacterium can convert 5-Cl PZA and PZA to their respective carboxylates at a low rate (Table 2). Observation of this activity is in accord with the recent report that mutant M. bovis strains deficient for de novo NAD biosynthesis can synthesize this essential cofactor from nicotinamide using the NAD salvage pathway (20). As this pathway is initiated by the deamidation of nicotinamide to nicotinic acid, it is likely that an as yet uncharacterized amidase(s) is expressed by these bacilli. Indeed, a BLAST-mediated search of the tubercle bacillus genome sequences revealed six uncharacterized pzaA homologs. The corresponding genes of M. tuberculosis strain H37Rv include (percent identity between PzaA and predicted protein homolog) Rv1263 (30%), Rv2363 (35%), Rv2888c (29%), Rv3011c (33%), Rv3175 (44%), and Rv3375 (35%). Although, this basal level of amidase activity was sufficient for utilization of nicotinamide by M. bovis, it was not robust enough to modulate susceptibility to PZA.
DISCUSSION
In this study, we demonstrate that ectopic expression of the M. smegmatis amidase PzaA results in concomitant resistance to 5-Cl PZA and susceptibility to PZA in M. smegmatis, M. bovis, and M. tuberculosis. In accord with previous observations on the effects of PZA and its derivatives on various species of mycobacteria (14, 26, 27), acquired susceptibility to PZA in M. smegmatis correlated with FAS I inhibition. This inhibition was rapid, dose dependent, and indistinguishable from the inhibition observed with 5-Cl PZA (26, 27). Thus, PZA and analogs, such as 5-Cl PZA, likely affect the same essential cellular function in different species of mycobacteria.
In contrast to that which was observed with PzaA, ectopic expression of the M. tuberculosis amidase PncA led to enhanced PZA susceptibility in M. bovis and M. tuberculosis yet had no notable effect on 5-Cl PZA turnover or susceptibility. Thus, PncA does not appear to recognize 5-Cl PZA as a substrate, consistent with the narrow substrate range of this amidase (2). Interestingly, M. bovis, which naturally lacks PncA activity, was found to slowly hydrolyze both PZA and 5-Cl PZA by an uncharacterized amidase activity. The presence of this cryptic amidase activity potentially explains the observation that M. bovis can utilize exogenous nicotinamide when the metabolite is present at high concentrations (20). Our results indicate that increased expression of such a broad substrate amidase represents a potential mechanism for 5-Cl PZA resistance. However, we predict that such a gain of function will also confer enhanced susceptibility to PZA.
The PzaA-mediated reciprocal resistance and susceptibility profiles are in accord with previous studies that showed that 5-Cl PA is a less potent antimycobacterial agent than 5-Cl PZA (4) and that elevated levels of PZase activity enhance PZA turnover and susceptibility in tubercle bacilli (2). The basis for the decreased activity of 5-Cl PA relative to 5-Cl PZA remains unresolved yet may be related to diminished uptake of 5-Cl PA due to the presence of its charged carboxylate group and/or decreased affinity of the compound for FAS I. In the case of PA, acidic pH has been suggested to enhance cellular accumulation by increasing the concentration of its conjugate acid (pKa, 2.9), which should diffuse more readily into the cell (24). The decreased antimycobacterial activity of 5-Cl PA relative to PA is consistent with a model for diminished uptake, as 5-Cl PA has a lower pKa, which dictates that the concentration of the 5-Cl PA conjugate acid will be lower than that for PA under equivalent pH conditions.
Previous studies have indicated that, in addition to PA formation and acidic pH of the local environment, PZA susceptibility of tubercle bacilli is dependent upon diminished cellular efflux relative to that of some nontubercle mycobacteria (24). Previous studies have shown that the rate of PZA deamidation affects the degree of susceptibility to the compound (2, 10). Here, we demonstrate that in M. smegmatis moderate deamidation of 5-Cl PZA was sufficient to confer 5-Cl PZA resistance, while PZA susceptibility was observed only with rapid PA formation. The latter indicates that there is a threshold of PA production that is necessary for PZA susceptibility, as has been suggested previously (23). It is likely that in our studies the PzaA-mediated PA formation exceeded the capacity of the M. smegmatis efflux system and enabled greater PA accumulation. Interestingly, a significant increase in PZA turnover markedly diminished the necessity for either acidic pH or impaired efflux for PZA susceptibility in tubercle bacilli. For M. tuberculosis, which was shown to be defective for efficient efflux of PA (24), a 12-fold increase in the rate of PZA deamidation by ectopic expression of PncA was sufficient to lower the MIC at neutral pH from 1,000 μg/ml to 62.5 μg/ml. Similar results were obtained for M. bovis, where the MIC was lowered from an undeterminable level to 62.5 μg/ml.
Thus far, we have been unable to isolate tubercle bacilli with spontaneous resistance to 5-Cl PZA, nor have we been able to isolate PzaA-independent spontaneously 5-Cl PZA-resistant strains of M. smegmatis. It is possible that mutations that lead to alterations in the target or transporter of this agent are extremely rare due to functional constraints. These results are in accord with previous reports that the occurrence of PA-resistant mutants of tubercle bacilli is beyond the limit of detection (17, 26). For 5-Cl PZA, we anticipate that resistance in tubercle bacilli can be mediated by mutations that affect the substrate specificity of PncA or by mutations that lead to increased expression of the cryptic amidase activity that we have described. It is expected that such a 5-Cl PZA-resistant mutant would show enhanced susceptibility to PZA due to increased formation of PA. Thus, the main finding of this study is that PZA and 5-Cl PZA form a complementary “resistance-proof” class of agents for M. tuberculosis therapy. PZA analogs with an ability to preclude resistance when used in combination show great promise in the global effort to eradicate TB.
Acknowledgments
We are grateful to David Thaler for helpful discussions regarding the manuscript.
This work was supported by a grant from NIAID (5R37AI026170). A.D.B. was a Merck Fellow of the Helen Hay Whitney Foundation.
Footnotes
Published ahead of print on 27 September 2010.
REFERENCES
- 1.Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 2.Boshoff, H. I. M., and V. Mizrahi. 2000. Expression of Mycobacterium smegmatis pyrazinamidase in Mycobacterium tuberculosis confers hypersensitivity to pyrazinamide and related amides. J. Bacteriol. 182:5479-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer, H. W. and Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 4.Cynamon, M. H., R. J. Speirs, and J. T. Welch. 1998. In vitro antimycobacterial activity of 5-chloropyrazinamide. Antimicrob. Agents Chemother. 42:462-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dye, C., S. Scheele, P. Dolin, V. Pathania, M. C. Raviglione, and for the WHO Global Surveillance and Monitoring Project. 1999. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 6.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox, W., and D. A. Mitchison. 1975. Short-course chemotherapy for pulmonary tuberculosis. Am. Rev. Respir. Dis. 111:325-353. [DOI] [PubMed] [Google Scholar]
- 8.Gandhi, N. R., A. Moll, A. W. Sturm, R. Pawinski, T. Govender, U. Lalloo, K. Zeller, J. Andrews, and G. Friedland. 2006. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368:1575-1580. [DOI] [PubMed] [Google Scholar]
- 9.Glover, R. T., J. Kriakov, S. J. Garforth, A. D. Baughn, and W. R. Jacobs, Jr. 2007. The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J. Bacteriol. 189:5495-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, M., Z. Sun, and Y. Zhang. 2000. Mycobacterium smegmatis has two pyrazinamidase enzymes, PncA and PzaA. J. Bacteriol. 182:3881-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konno, K., F. M. Feldmann, and W. McDermott. 1967. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am. Rev. Respir. Dis. 95:461-469. [DOI] [PubMed] [Google Scholar]
- 12.Lee, S., J. Kriakov, C. Vilcheze, Z. Dai, G. F. Hatfull, and W. R. Jacobs. 2004. Bxz1, a new generalized transducing phage for mycobacteria. FEMS Microbiol. Lett. 241:271-276. [DOI] [PubMed] [Google Scholar]
- 13.McDermott, W., and R. Tompsett. 1954. Activation of pyrazinamide and nicotinamide in acidic environments in vitro. Am. Rev. Tuberc. 70:748-754. [DOI] [PubMed] [Google Scholar]
- 14.Ngo, S. C., O. Zimhony, W. J. Chung, H. Sayahi, W. R. Jacobs, Jr., and J. T. Welch. 2007. Inhibition of isolated Mycobacterium tuberculosis fatty acid synthase I by pyrazinamide analogs. Antimicrob. Agents Chemother. 51:2430-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raynaud, C., M. A. Laneelle, R. H. Senaratne, P. Draper, G. Laneelle, and M. Daffe. 1999. Mechanisms of pyrazinamide resistance in mycobacteria: importance of lack of uptake in addition to lack of pyrazinamidase activity. Microbiology 145:1359-1367. [DOI] [PubMed] [Google Scholar]
- 16.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662-667. [DOI] [PubMed] [Google Scholar]
- 18.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 19.Sun, Z., and Y. Zhang. 1999. Reduced pyrazinamidase activity and the natural resistance of Mycobacterium kansasii to the antituberculosis drug pyrazinamide. Antimicrob. Agents Chemother. 43:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilchèze, C., B. Weinrick, K.-W. Wong, B. Chen, and W. R. Jacobs, Jr. 2010. NAD(+) auxotrophy is bacteriocidal for the tubercle bacilli. Mol. Microbiol. doi: 10.1111/j.1365-2958.2010.07099.x. [DOI] [PMC free article] [PubMed]
- 21.Wayne, L. G. 1974. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am. Rev. Respir. Dis. 109:147-151. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, Y., and D. Mitchison. 2003. The curious characteristics of pyrazinamide: a review. Int. J. Tuberc. Lung Dis. 7:6-21. [PubMed] [Google Scholar]
- 23.Zhang, Y., S. Permar, and Z. Sun. 2002. Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J. Med. Microbiol. 51:42-49. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, Y., A. Scorpio, H. Nikaido, and Z. Sun. 1999. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Bacteriol. 181:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, Y., M. M. Wade, A. Scorpio, H. Zhang, and Z. Sun. 2003. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J. Antimicrob. Chemother. 52:790-795. [DOI] [PubMed] [Google Scholar]
- 26.Zimhony, O., J. S. Cox, J. T. Welch, C. Vilchèze, and W. R. J. Jacobs. 2000. Pyrazinamide inhibits the eukaryotic-like fatty acid synthetase I (FASI) of Mycobacterium tuberculosis. Nat. Med. 6:1043-1047. [DOI] [PubMed] [Google Scholar]
- 27.Zimhony, O., C. Vilcheze, M. Arai, J. T. Welch, and W. R. Jacobs, Jr. 2007. Pyrazinoic acid and its n-propyl ester inhibit fatty acid synthase type I in replicating tubercle bacilli. Antimicrob. Agents Chemother. 51:752-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimhony, O., C. Vilchèze, and W. R. Jacobs, Jr. 2004. Characterization of Mycobacterium smegmatis expressing the Mycobacterium tuberculosis fatty acid synthase I (fas1) gene. J. Bacteriol. 186:4051-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]