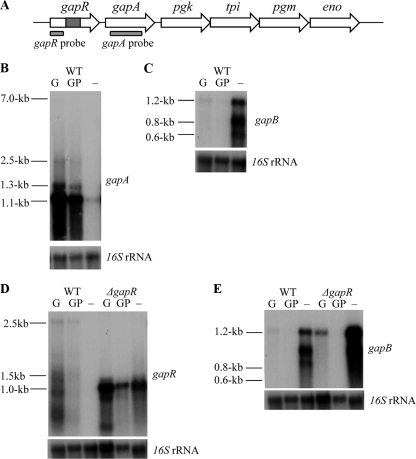
FIG. 4.
(A) Schematic representation of the glycolytic operon with a ΔgapR mutation, indicating the position of the gapR and gapA probes used for Northern blot analysis. Genes are putative operon regulator (gapR), glyceraldehyde-3-phosphate dehydrogenase (gapA), phosphoglycerate kinase (pgk), triphosphate isomerase (tpi), phosphoglycerate mutase (pgm), and enolase (eno). Northern blot analysis is shown of gapA transcript (B) and gapB transcript (C) expression in response to glucose induction in strain 8325-4 and gapR transcript (D) and gapB transcript (E) expression in response to glucose induction in both wild-type (WT) 8325-4 and 8325-4 ΔgapR strains. Total RNA was extracted from cells grown for 5 h in TM broth with 1% glucose added at time zero (G), 1% glucose added 1 h before cells were harvested (GP), and without glucose (−). Gels presented are representative of experiments that were repeated two times using RNA extracts from cultures grown on different days, with similar results observed each time. Blots were stripped and rehybridized with a control probe (16S) to ensure equal loading of RNA in each case.