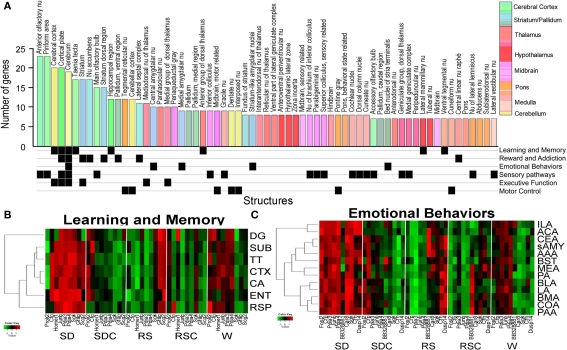
Figure 6.
Signatures of sleep deprivation by anatomic region. (A) The histogram shows the number of genes exhibiting differential expression between SD and SDC (ANOVA p < 0.05) per anatomic region based upon automated quantification of triplicate ISH data from 53 genes using expression energy. Histogram bars are color-coded by the anatomic ontology based upon the Allen Reference Atlas for adult mouse brain, and the key to the colors is shown to the far right. Below the histogram, anatomic regions are annotated for involvement in different functional pathways based on the published literature. (B,C) Heatmaps showing the response of genes to five behavioral conditions for subsets of brain regions (y-axis) involved in either learning and memory (B) or emotional behaviors (C). Along the x-axis, genes which exhibited differential gene expression across five conditions for any of the given brain regions as determined by triplicate ISH (ANOVA, p < 0.05) are shown. Abbreviations: DG, dentate gyrus; SUB, subiculum; TT, tania tecta; CTX, cerebral cortex; CA, Ammon's Horn CA region; ENT, entorhinal cortex; RSP, retrosplenial cortex; ILA, infralimbic area; ACA, anterior cingulate area; CEA, central nucleus of amygdala; sAMY, striatum-like amygdalar nuclei; AAA, anterior amygdalar area; BST, bed nuclei of stria terminalis; MEA, medial amygdalar nucleus; PA, posterior amygdalar nucleus; BLA, basolateral amygdala; LA, lateral amygdala; BMA, basomedial amygdala; COA, cortical amygdala; PAA, piriform-amygdalar area.