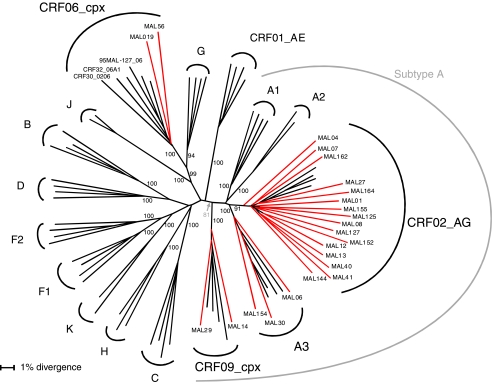
FIG. 1.
Phylogenetic classification of 23 Malian HIV-1 isolates based on the full-length env region. A representative sequence from each Malian patient (highlighted in red) and 116 reference subtypes and available CRFs (at least two representative sequences of subtypes, subsubtypes, and of CRF01 to CRF15, CRF18, CRF19 as well as a prototype sequence from CRF16, CRF17, CRF20–CRF25, CRF28–CRF33, and CRF35–CRF37) were initially used to construct the tree; some references have been omitted here for clarity. For a complete list of reference sequences used in the analysis, see Table 2. A phylogenetic tree was constructed by the neighbor-joining method with the HKY85 model of evolution. Bootstrap percentile values from 1000 replications are shown at nodes defining major grouping of sequences. Clustering of subtype/subsubtype sequences are delineated in the figure. Previously generated sequences from Mali, 95ML127 (CRF06), were included in the analysis. Recently classified CRF30_020611 and CRF32_06A1,31 both containing CRF06_cpx segments in their env region, were also included in the analysis and found within the CRF06_cpx radiation with a high bootstrap value of 100%.