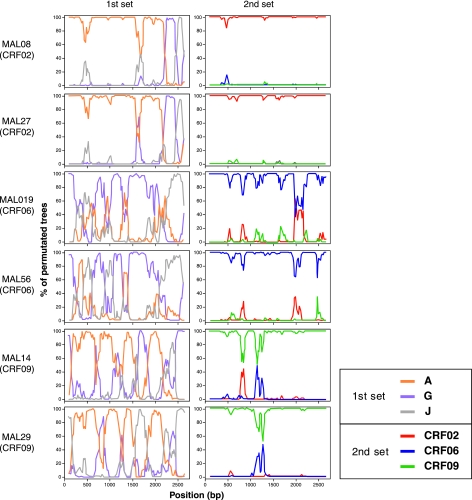
FIG. 2.
Bootscanning analyses of 2.6-kb env regions of HIV-1 sequences of CRF02 (MAL08 and 27), CRF06 (MAL019 and 56), and CRF09 (MAL14 and 29). Two sets of bootscanning were performed: The first set of bootscanning plots depicts relationship of the Mali sequences to the representative strains of HIV-1 nonrecombinant subtypes (A: 94SE_Q23, G: DRCBL, and J: 93SE_7887). The second set of bootscanning plots delineates relationship of the Mali sequences to the prototype strain of CRF02 (IbNG), CRF06 (BFP90), and CRF09 (96GH2911). For the bootscanning plots, the SimPlot software performed bootscanning on neighbor-joining trees by using SEQBOOT (100 replicates), DNADIST (with the Kimura's two-parameter method and a transition/transversion ratio of 2.0), and CONSENSE from the PHYLIP package,32 for a 200 bp window moving along the alignment with increments of 20 bp.