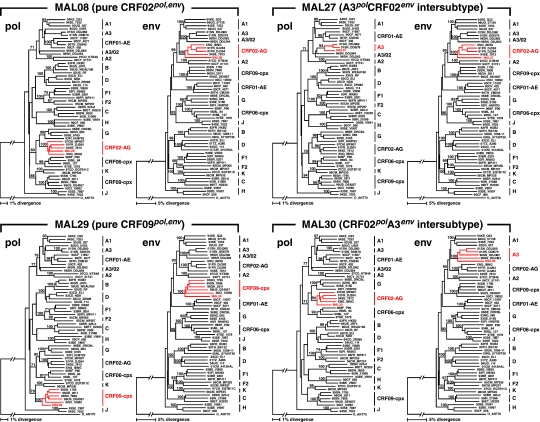
FIG. 3.
Subregion tree analysis of the pol and full-length env sequences. Four representative patterns are shown. Each gene segment was subjected to separate phylogenetic analyses based on the neighbor-joining method to identify the subtype or circulating recombinant form (CRF) origin of the segment. The stability of the nodes was assessed by the use of bootstrap resampling of 1000 replications. Bootstrap values above 70% are shown. The clusters for subtype/subsubtypes and CRF01_AE, CRF02_AG, CRF06_cpx, and CRF09_cpx are outlined with a vertical bar on the right of each tree. The genotype assignments of each Mali strain in the respective phylogenetic analysis are highlighted in red in each tree. All the HIV-1 reference sequences were retrieved from the HIV sequence database, Los Alamos National Library. For a complete list of reference sequences used in the analysis, see Table 3.