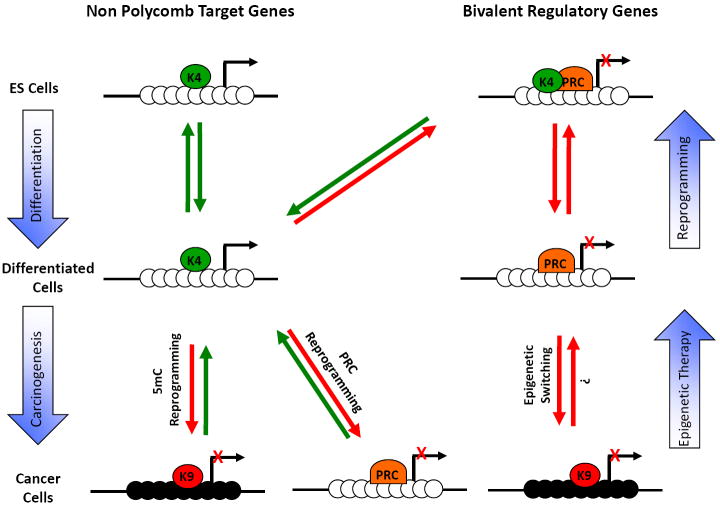
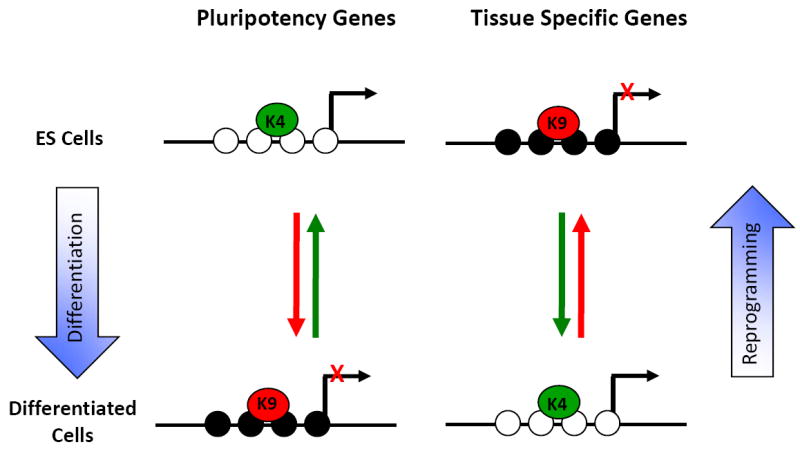
Figure 3. Epigenome landscaping during differentiation and carcinogenesis.
(a) CpG rich regions: In embryonic stem cells, most CpG rich regions contained within promoters are marked by H3K4me3 and can be subdivided in two groups. The first group comprises genes that are not Polycomb targets and are usually expressed in ES cells. The second group comprises Polycomb target genes. This group adopts a bivalent chromatin structure and is usually repressed in ES cells. During cell differentiation, the bivalent chromatin structure becomes monovalent and some genes retain only the H3K4me3 mark (K4) while others retain only the H3K27me3 mark (PRC). Interestingly, during reprogramming towards pluripotency, most of these genes reacquire the bivalent structure [73, 74]. During carcinogenesis, a group of expressed genes become de-novo repressed, either by DNA methylation (5mC reprogramming) or by histone methylation (PRC reprogramming). In addition, another group of genes already repressed by Polycomb gains a more stable repression by DNA methylation (epigenetic switching) [85]. The gain of DNA methylation is usually accompanied by H3K9 methylation (K9). Black circles are methylated CpG sites and white circles are unmethylated.
(b) CpG poor regions: In ES cells, pluripotency genes located in non-CGI are marked by H3K4me3, the CpG sites are unmethylated and the genes are expressed. During cell differentiation, these genes gain DNA methylation, the repressive H3K9me3 mark and are not expressed. Tissue specific genes are silenced by DNA methylation in ES cells and during differentiation these genes are expressed in some tissues, while remaining silenced in others. During reprogramming, this group can revert their chromatin structure, becoming very similar to that of ES cells [70]. Black circles are methylated CpG sites and white circles are unmethylated.