Abstract
Background: DNA strand breaks pose the greatest threat to genomic stability. Genetically determined mutagen sensitivity predisposes individuals to a variety of cancers, including glioma. However, polymorphisms in DNA strand break repair genes that may determine mutagen sensitivity are not well studied in cancer risk, especially in gliomas.
Methods: We correlated genotype data for tag single-nucleotide polymorphisms (tSNPs) of DNA strand break repair genes with a gamma-radiation-induced mutagen sensitivity phenotype [expressed as mean breaks per cell (B/C)] in samples from 426 glioma patients. We also conducted analysis to assess joint and haplotype effects of single-nucleotide polymorphisms (SNPs) on mutagen sensitivity. We further validate our results in an independent external control group totaling 662 subjects.
Results: Of the 392 tSNPs examined, we found that mutagen sensitivity was modified by one tSNP in the EME2 gene and six tSNPs in the RAD51L1 gene (P < 0.01). Among the six RAD51L1 SNPs tested in the validation set, one (RAD51L1 rs2180611) was significantly associated with mutagen sensitivity (P = 0.025). Moreover, we found a significant dose–response relationship between the mutagen sensitivity and the number of adverse tSNP genotypes. Furthermore, haplotype analysis revealed that RAD51L1 haplotypes F-A (zero adverse allele) and F-E (six adverse alleles) exhibited the lowest (0.42) and highest (0.93) mean B/C values, respectively. A similar dose–response relationship also existed between the mutagen sensitivity and the number of adverse haplotypes.
Conclusion: These results suggest that polymorphisms in and haplotypes of the RAD51L1 gene, which is involved in the double-strand break repair pathway, modulate gamma-radiation-induced mutagen sensitivity.
Introduction
The genetic instability driving tumorigenesis is fueled by DNA damage and by errors made by the DNA machinery (1). Of all the DNA errors, strand breaks are perhaps the most dangerous and major threats to the genomic stability in humans. Both single-strand breaks and double-strand breaks (DSBs) can arise from ionizing radiation or X-rays, free radicals and chemicals. Single-strand breaks and damaged bases are repaired by the action of enzymes in the base excision repair (BER) pathway (1). These lesions may or may not impede transcription and replication, although they frequently cause miscoding errors. BER is therefore particularly relevant for preventing mutagenesis. However, DSBs are more problematic because both strands are affected. To repair such breaks, human cells employ either an error-prone non-homologous end-joining mechanism, which cements broken DNA ends together regardless of their genomic position or identity, or an error-free mechanism that recreates the unbroken DNA sequence by homologous recombination with an undamaged homologous strand (1). Homologous recombination repair tends to occur in dividing cells during the S phase of the cell cycle, when the freshly replicated sister chromatid offers a perfect template for exchange; this process is therefore essential for the maintenance of genome stability and tumor avoidance.
It is believed that deficient DNA strand repair results in an abnormally high frequency of chromosomal aberrations after exposure to radiation and that this process may underlie the mutagen sensitivity associated with susceptibility to cancer. The rationale behind these associations between genes and cancer risk is that these radiation-sensitive genetic variants may result in alterations in DNA repair capacity (DRC), an intermediate phenotype. However, correlations between genotype and phenotype have not been explored for the majority of these polymorphisms in DNA strand break repair pathway [including BER and double-strand break repair (DSBR)] genes in risk of cancer, especially in glioma.
The in vitro mutagen sensitivity is a phenotypic measure of genetic susceptibility to various forms of cancer. It is measured by quantifying the number of chromatid breaks induced by known mutagens in short-term-cultured lymphocytes and has been used as an indirect measure of an individual's DRC that serves as an intermediate phenotype for cancer risk (2). Epidemiologic studies have suggested that mutagen sensitivity is a predisposing factor for a variety of cancers, including glioma (3). In previous studies (3,4), we found that cells from glioma patients were more sensitive to gamma-radiation than that from normal control subjects. To substantiate our preliminary findings, we have expanded the study to include 426 glioma patients with gamma-radiation-induced mutagen sensitivity phenotype. We hypothesized that variants of DNA strand break repair genes may influence mutagen sensitivity phenotype, and alterations in these genes could cause altered mutagen sensitivity that modulate glioma risk. In the present study, we used a pathway-based approach to investigate 392 tag single-nucleotide polymorphisms (tSNPs) from 45 genes involved in DNA strand break repair pathways, including BER and DSBR. Lastly, we tested our findings in an additional independent control group.
Materials and methods
Study subjects
The population for this study was a subset of patients from a prospective study of malignant glioma patients consecutively diagnosed and treated at The University of Texas M. D. Anderson Cancer Center, Houston, TX (5,6). The patients had histologically confirmed and previously untreated malignant gliomas and were participants in an ongoing epidemiological study of glioma from 1992. The study recruited participants sequentially but excluded those having previously received chemotherapy or radiotherapy. Only Caucasian patients were included. Non-Caucasian patients (11 Hispanic, 12 African-American and 3 Asian) were excluded since the number of patients was too small for a statistically meaningful analysis. Other exclusion criteria included previous diagnosis of other cancers and previous radiation treatment for other illnesses. The study was approved by M. D. Anderson IRB, and written informed consent was obtained from each participant.
A questionnaire administered to the patients by trained interviewers yielded comprehensive data on their history of personal health, family history of brain tumors (first-degree), occupation, exposures, smoking and alcohol consumption. A 20 ml blood specimen was obtained from all patients.
Mutagen sensitivity assay
Gamma-radiation was selected as the test mutagen because it induces both single-strand breaks and DSBs and because radiation exposure is the only documented risk factor for brain tumors (7,8). The assay has been described in detail elsewhere (3,4). Briefly, we used a previously established optimal dose of gamma-radiation (1.5 Gy) to investigate the mutagen sensitivity of peripheral blood lymphocytes. This dose is within the range of the daily dose generally used in clinical practice and is also similar to the amount of radiation therapy, most patients undergoing radiation therapy receives as a single daily dose (1.2–2 Gy per fraction). Two standard lymphocyte cultures were established from each subject in blood samples as described elsewhere (2). Fifty metaphases/samples were scored and only frank chromatid breaks were recorded; chromatid gaps or attenuated regions were discarded. The average number of chromatid breaks per cell (B/C) was then calculated.
Selection of tSNPs and genotyping assays
We identified a set of 17 genes involved in the BER pathway and 28 genes involved in the DSBR pathway by selecting all the genes listed in the Human DNA repair genes reviewed by Wood et al. (9) (http://sciencepark.mdanderson.org/labs/wood/DNA_Repair_Genes.html) and then adding five genes listed in the PANTHER database (http://www.pantherdb.org/pathway/) as being related to the DSBR pathway (ATM, RTEL1, BTBD2, HMGA2 and XRCC9). A total of 718 single-nucleotide polymorphisms (SNPs) were identified on the above 50 genes from Human 610-Quad Bead Chip (Illumina, San Diego, CA). Some SNPs on the chip were in linkage disequilibrium (LD) with each other. To select the LD tSNPs, we analyzed the genotype data with the HAPLOVIEW software (http://www.broad.mit.edu/mpg) using the tag procedure with a threshold of r2 >0.8 and a minor allele frequency >0.05. We selected a total of 308 LD tSNPs; the vast majority of which were located in intronic regions (see supplementary Table 1, is available at Carcinogenesis Online, for the list of the tSNPs).
We contracted with Illumina to conduct the genotyping using the Illumina Infinium HD Human 610-Quad Bead Chips according to the manufacturer's protocols (Illumina).
Statistical methods
Descriptive statistics [mean, median, range and standard deviation (SD)] of mutagen sensitivity were computed for all participants and for subgroups by age, sex, first-degree family history of brain tumor and histological type of tumors. SNP variables were divided into two categories according to their best genetic model as selected by Akaike's information criterion (10).
Mutagen sensitivity by genotype was compared using Student's t-test or analysis of variance. Mutagen sensitivity was also dichotomized at the median to calculate odds ratios (ORs) associated with each genotype: B/C values greater than the median were considered to be mutagen sensitive, whereas values less than the median were considered to be non-sensitive. ORs and 95% confidence intervals (CIs) were calculated by unconditional logistic regression analysis with adjustment for age and sex. The joint effect analysis was evaluated by adding up the number of adverse alleles of the significant SNPs identified from the main effects analysis. Adverse alleles were defined as the minor allele of the risk SNPs and the common allele of the protective SNPs.
Pairwise LD was examined using Lewontin's standardized coefficient D′ (11). Haplotype block structures were defined by the method of LD CIs method (12). The HAPLO.STATS package (http://www.mayo.edu/hsr/Sfunc.html) in the software language R was used for the haplotype analysis (13). Multiple linear regression models with Bonferroni correction were fit including the probabilities of the common haplotypes (frequency > 0.05) to compare mutagen sensitivity levels among the haplotypes in each block and to estimate mutagen sensitivity B/C values (mean ± SD). We used a Dirichlet distribution to calculate the correlation between each predictor variable in a fitted model to verify that variables were not correlated (14). The mutagen-sensitive and non-sensitive groups were also compared using a logistic regression analysis to evaluate the association between each haplotype and sensitivity. Empirical P values, based on 10 000 simulations, were computed for the global score test and each of the haplotype-specific score tests. All statistical tests were two sided, and a P value <0.05 was considered statistically significant.
To evaluate the chance of obtaining a false-positive association in our data set, we used the false-positive report probability (FPRP) test (15). For our analyses, we used the moderate range of prior probabilities 0.05 and FPRP cutoff value of 0.2 as suggested by the authors for summary analyses.
Subjects for the replication phase
As a validation group, we obtained data for 662 healthy controls from another independent population (16,17). Controls were Caucasian people without prior cancer history (except for non-melanoma skin cancer) who were identified through Kelsey-Seybold Clinics, the largest multispecialty physician group in the Houston metropolitan area.
The mean age was 62.9 for this control group. There were 519 men (78.4%) and 143 women (21.6%). Mutagen sensitivity assay was treated with 1.25 Gy of gamma-radiation delivered from a cesium-137 irradiator. The number of chromatid breaks in 50 metaphases per sample was counted and expressed as the average number of B/C. Replication genotyping of the six SNPs was conducted using the Illumina 610k chip at M.D. Anderson from another study (17).
Results
Patient and sample characteristics
Table I summarizes the characteristics of the 426 glioma cases included in this analysis. The mean age at diagnosis was 44.9 years (median, 45.5 years; SD, 12.4 years). There were 267 men (62.7%) and 159 women (37.3%). The mean B/C was 0.62, with a median value of 0.48 (SD, 0.50) and a range of 0.12–3.82. Any individual who expressed a value higher than the median (>0.48 B/C) was considered mutagen sensitive.
Table I.
Radiation-induced mutagen sensitivity status by select variables in glioma patients
| Variables | All cases (N = 426) |
Comparisona |
||||
| N (%)b | B/C mean (SD) | P-value | Non-sensitive (n = 223) | Sensitive (n = 203) | P-value | |
| Age, years | ||||||
| Mean | 44.9 | |||||
| Range | 18–78 | |||||
| ≤45 | 213 (50.0) | 0.66 (0.54) | 0.15 | 110 (49.3) | 103 (50.7) | 0.85 |
| >45 | 213 (50.0) | 0.59 (0.46) | 113 (50.7) | 100 (49.3) | ||
| Sex | ||||||
| Male | 267 (62.7) | 0.60 (0.45) | 0.30 | 148 (66.4) | 119 (58.6) | 0.08 |
| Female | 159 (37.3) | 0.66 (0.57) | 75 (33.7) | 84 (41.4) | ||
| Smoke | ||||||
| Ever | 186 (43.7) | 0.63 (0.51) | 0.91 | 96 (47.1) | 90 (48.9) | 0.72 |
| Never | 202 (47.4) | 0.62 (0.52) | 108 (52.9) | 94 (51.1) | ||
| First-degree family history of brain tumor | ||||||
| Yes | 13 (3.1) | 0.38 (0.13) | 0.16 | 10 (4.7) | 3 (1.6) | 0.07 |
| No | 394 (92.5) | 0.63 (0.49) | 205 (95.3) | 189 (98.4) | ||
| Histologyc | ||||||
| High grade | 214 (50.3) | 0.58 (0.40) | 0.02d | 114 (51.1) | 100 (49.3) | 0.71 |
| Medium grade | 67 (15.7) | 0.78 (0.70) | 32 (14.3) | 35 (17.2) | ||
| Low grade | 145 (34.0) | 0.62 (0.51) | 77 (34.5) | 68 (33.5) | ||
Comparison dichotomized at the median of B/C (0.48) among 426 study subjects.
Numbers do not add up to the column totals due to missing values.
High-grade glioma (glioblastoma), medium-grade glioma (anaplastic astrocytoma) and low-grade glioma (oligodendroglioma, not-otherwise-specified astrocytoma and mixed glioma).
Bold numbers indicate significant values at P <0.05.
We found statistically significant differences in mutagen sensitivity by glioma histology. The mean B/C was 0.58 for the 214 glioblastoma multiformes, 0.78 for the 67 anaplastic astrocytomas and 0.62 for the other 145 gliomas (oligodendrogliomas or mixed glioma). All the other variables, including age, sex and first-degree family history of brain tumor, did not influence the gamma-radiation-induced mutagen sensitivity. However, after dividing the samples into two groups based on mutagen sensitivity (sensitive versus non-sensitive) using the median B/C value (0.48), we found a marginally significant difference in mutagen sensitivity by gender (P = 0.08) and first-degree family history of brain tumor (P = 0.07).
Association between mutagen sensitivity and genetic variants
Individual SNP association analysis.
Of the 392 tSNPs analyzed, 31 were significantly associated with mutagen sensitivity (P ≤ 0.05), and seven of these had statistically significant genotype–phenotype correlations in individual analyses (P ≤ 0.01; one in EME2 and six in RAD51L1; Table II). Compared with wild-type homozygous and heterozygous patients, individuals with homozygous variants for the three risk SNPs (recessive model: RAD51L1 rs2256608, rs2064827 and rs4902623) exhibited significantly higher radiation-induced B/C values (P = 0.004, 0.004 and 0.002, respectively). Compared with individuals with the wild-type homozygous phenotype, those with the variant-type homozygous and heterozygous protective SNPs (dominant model: RAD51L1 rs12432197, rs12893578 and rs2180611) exhibited significantly lower B/C values (P = 0.0007, 0.00008 and 0.002, respectively).
Table II.
Association between strand break repair pathway gene SNP genotype and mutagen sensitivity
| Gene | SNP ID | Genic position | Predicted functiona | MAF | Genotype | All cases (N = 426) |
Comparisonb |
|||||
| n (%) | B/C mean (SD) | P | Non-sensitive/sensitive | OR (95% CI) | P | FPRPc | ||||||
| Risk effect | ||||||||||||
| RAD51L1 | rs2256608 | Intron 12 | Recombination hot spot | 0.211 | AG/GGR | 404 (94.84) | 0.61 (0.49) | 0.004 | 216/188 | Reference | 0.038 | |
| AA | 22 (5.16) | 0.92 (0.64) | 7/15 | 2.53 (1.01–6.37) | 0.750 | |||||||
| RAD51L1 | rs2064827 | 3′ UTR | TFBS | 0.225 | TT/TGR | 400 (93.89) | 0.61 (0.49) | 0.004 | 214/186 | Reference | 0.047 | |
| GG | 26 (6.11) | 0.90 (0.63) | 9/17 | 2.20 (0.97–5.05) | 0.744 | |||||||
| RAD51L1 | rs4902623 | 3′ UTR | TFBS | 0.435 | AA/AGR | 356 (83.57) | 0.59 (0.45) | 0.002 | 196/160 | Reference | 0.008 | |
| GG | 70 (16.43) | 0.79 (0.69) | 27/43 | 1.92 (1.13–3.22) | 0.312 | |||||||
| EME2 | rs2437732 | 3′ UTR | microRNA target site | 0.396 | AC/AAR | 368 (86.39) | 0.60 (0.47) | 0.007 | 201/167 | Reference | 0.013 | |
| CC | 58 (13.61) | 0.79 (0.65) | 22/36 | 1.96 (1.11–3.46) | 0.422 | |||||||
| Protective effect | ||||||||||||
| RAD51L1 | rs12432197 | 3′ UTR | TFBS | 0.473 | TT | 116 (27.23) | 0.76 (0.67) | 0.0007 | 51/65 | Reference | 0.022 | |
| TC/CCD | 310 (72.77) | 0.57 (0.41) | 172/138 | 0.62 (0.41–0.96) | 0.423 | |||||||
| RAD51L1 | rs12893578 | 3′ UTR | TFBS | 0.424 | AA | 129 (30.28) | 0.77 (0.65) | <0.0001 | 53/76 | Reference | 0.001 | |
| AG/GGD | 297 (69.72) | 0.56 (0.41) | 170/127 | 0.52 (0.34–0.79) | 0.067 | |||||||
| RAD51L1 | rs2180611 | 3′ UTR | TFBS | 0.380 | GG | 153 (35.92) | 0.72 (0.58) | 0.002 | 62/91 | Reference | 0.0002 | |
| AG/AAD | 273 (64.08) | 0.57 (0.44) | 161/112 | 0.46 (0.31–0.70) | 0.016 | |||||||
| Joint effect | ||||||||||||
| No. of adverse alleles | 0 | 199 (46.7) | 0.55 (0.44) | <0.0001 | 124/75 | Reference | <0.0001 | |||||
| 1–2 | 117 (27.5) | 0.58 (0.33) | 58/59 | 1.72 (1.08–2.73) | ||||||||
| 3–4 | 96 (22.5) | 0.73 (0.61) | 37/59 | 2.67 (1.61–4.41) | ||||||||
| 5–7 | 14 (3.3) | 1.30 (1.01) | 4/10 | 3.94 (1.19–13.05) | ||||||||
Akaike's information criterion was used to determine the genetic model for each SNP. D, dominant genetic model; R, recessive genetic model.
Functional predictions based on the in silico tool SNP function portal (http://brainarray.mbni.med.umich.edu/Brainarray/Database/SearchSNP/snpfunc.aspx) (18).
Dichotomized at the median of B/C (0.48), analysis adjusted by age and sex.
Bold data have a noteworthy association at prior 0.05, FPRP ≤0.2.
We treated the minor allele of the four risk-effect SNPs and the common allele of the three protective-effect SNPs as the adverse alleles and set individuals with 0 adverse alleles as the reference group. Adverse genotypes—for risk SNPs: RAD51L1 rs2256608 AA, rs2064827 GG and rs4902623 GG and EME2 rs2437732 CC; for protective SNPs: RAD51L1 rs12432197 TT, rs12893578 AA and rs2180611 GG.
Among the other DSBR genes, significant differences in mutagen sensitivity were observed for EME2 rs2437732. Individuals homozygous for EME2 rs2437732 variant (recessive) exhibited significantly higher gamma-radiation-induced B/C than individuals with the wild-type homozygous or heterozygous variants (P = 0.007). We did not observe any other SNPs in the same DSBR pathway with significant genotype–phenotype correlations. In addition, we did not find any associations between mutagen sensitivity and genetic polymorphisms in the BER genes, even when we used a P-value cutoff of 0.05.
We next assessed mutagen sensitivity as modified by the combined effects of the seven SNPs found to have a significant correlation with phenotype. We treated the minor allele of the four risk-effect SNPs (EME2 rs2437732 and RAD51L1 rs2256608, rs2064827 and rs4902623) and the common allele of the three protective-effect SNPs (RAD51L1 rs12432197, rs12893578 and rs2180611) as adverse alleles and set individuals with zero adverse alleles as the reference group. In this analysis, we observed that the mean B/C values increased progressively as the number of adverse allele increased: 0.55 for those carrying zero, 0.58 for those carrying one to two, 0.73 for those carrying three to four and 1.30 for those carrying five to seven adverse alleles (P for trend < 0.0001). When we compared the mutagen-sensitive and non-sensitive subgroups with subjects carrying the zero adverse genotype, mutagen sensitivity increased by 1.72-fold (95% CI, 1.08–2.73) in individuals carrying one to two, 2.67-fold (95% CI, 1.61–4.41) in individuals carrying three to four and 3.94-fold (95% CI, 1.19–13.05) in individuals carrying five to seven adverse alleles (Table II).
To evaluate the robustness of these findings, we calculated FPRP values at 0.05 levels of prior probabilities for the seven SNPs. As shown in Table II, two of these seven SNPs (RAD51L1 rs12893578 and rs2180611) remained noteworthy (FPRP ≤ 0.2).
Haplotype block structure and LD analysis.
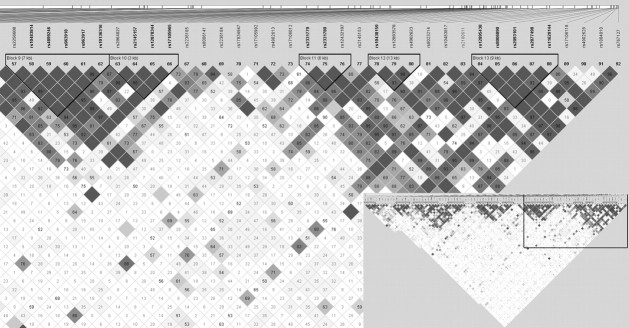
Because six tSNPs of the RAD51L1 gene showed a significant correlation with mutagen sensitivity, we compared mutagen sensitivity among the RAD51L1 haplotypes. Figure 1 shows a plot of the pairwise LD (D′) values for the 92 tSNPs and LD structures of RAD51L1. RAD51L1 (∼700 kb) is highly polymorphic; it has extensive haplotype diversity and weak LD across the entire gene. Thirteen blocks with high LD were identified: block 1, encompassing the majority of the gene from the upstream region intron 1 through intron 6; blocks 2–8, spanning intron 6–9; block 9, ranging from intron 9 to the 3′ untranslated region (UTR) and blocks 10–13 covering predominantly the 3′ UTR (Figure 1).
Fig. 1.
.LD plots and haplotype block structure of the RAD51L1. Haplotype blocks were defined according to the criteria reported by Gabriel et al. (12). The values indicate the LD relationship between each pair of SNPs; darker shading denotes a greater extent of LD between SNPs.
Because the significant tSNPs of RAD51L1 are located in block 9 (rs2256608), block 10 (rs2064827), block 11 (12432197), block 12 (rs12893578 and rs4902623) and the interblock region between blocks 12 and 13 (rs2180622), we looked at the association between the common haplotypes of blocks 9–12 in RAD51L1 and the mutagen sensitivity. Supplementary Table 2, is available at Carcinogenesis Online, shows the D′ and r2 between pairs of SNPs for each blocks in RAD54L1. As shown in Table III, one protective haplotype, 11-B (mean B/C = 0.47; adjusted OR, 0.66; 95% CI, 0.45–0.97) was identified in block 11; and two risk haplotypes were identified in block 12: 12-B (mean B/C = 0.61; adjusted OR, 1.51; 95% CI, 1.10–2.08) and 12-C (mean B/C = 0.71; adjusted OR, 1.62; 95% CI, 1.05–2.47). In further analysis, a global score test showed statistically significant differences in the haplotype profile for block 12 (global P = 0.042). For blocks 9 and 10, although no haplotypes were found to be significantly associated with mutagen sensitivity, both haplotypes 9-C (mean B/C = 0.74) and 10-C (mean B/C = 0.72), which carried a risk allele that was individually associated with increased mutagen sensitivity, conferred the highest mutagen sensitivity (B/C value) among the observed haplotypes.
Table III.
Association between haplotypes in blocks 9, 10, 11 and 12 of RAD51L1 and mutagen sensitivity
| Blocks | Haplotypea | All cases |
Comparisonb |
||||
| n (%) | Estimated B/C, mean (SD) | P-value | Percent non-sensitive/sensitive | OR (95% CI) | Global test, Pc | ||
| Block 9 | 0.13 | ||||||
| 9-A (GGAATT) | 124 (29.1) | 0.56 (0.61) | <0.0001 | 29.1/29.1 | Reference | ||
| 9-B (GGGACC) | 99 (23.3) | 0.55 (0.63) | 22.9/23.7 | 1.00 (0.69–1.44) | |||
| 9-C (AGAGCC) | 89 (21.1) | 0.74 (0.62) | 23.6/18.7 | 1.32 (0.93–1.90) | |||
| 9-D (GGAGCC) | 67 (15.7) | 0.40 (0.65) | 12.8/18.2 | 0.70 (0.46–0.06) | |||
| 9-E (GAAACT) | 41 (9.7) | 0.42 (0.68) | 10.3/9.0 | 1.21 (0.73–1.99) | |||
| Block 10 | 0.12 | ||||||
| 10-A (TTGT) | 122 (28.6) | 0.57 (0.62) | <0.0001 | 28.3/28.8 | Reference | ||
| 10-B (TCGT) | 105 (24.6) | 0.56 (0.63) | 24.6/24.6 | 1.07 (0.74–1.54) | |||
| 10-C (GTAT) | 95 (22.3) | 0.72 (0.62) | 24.4/20.5 | 1.27 (0.89–0.79) | |||
| 10-D (TCGC) | 62 (14.5) | 0.44 (0.67) | 12.1/16.7 | 0.74 (0.48–1.14) | |||
| 10-E (TTAT) | 42 (9.8) | 0.40 (0.68) | 10.6/9.1 | 1.23 (0.74–2.04) | |||
| Block 11 | 0.15 | ||||||
| 11-A (GGT) | 224 (52.7) | 0.57 (0.40) | <0.0001 | 554/50.2 | Reference | ||
| 11-B (GAC) | 84 (19.8) | 0.47 (0.66) | 17.0/22.3 | 0.66 (0.45–0.97) | |||
| 11-C (TGC) | 76 (17.9) | 0.52 (0.63) | 17.2/18.5 | 0.84 (0.58–1.20) | |||
| 11-D (GGC) | 41 (9.6) | 0.42 (0.69) | 10.3/9.0 | 1.05 (0.65–1.71) | |||
| Block 12 | 0.042 | ||||||
| 12-A (CGA) | 179 (42.1) | 0.44 (0.62) | <0.0001 | 37.6/46.1 | Reference | ||
| 12-B (CAG) | 159 (37.5) | 0.61 (0.60) | 40.8/34.4 | 1.51 (1.10–2.08) | |||
| 12-C (CAA) | 60 (14.1) | 0.71 (0.68) | 15.6/12.9 | 1.62 (1.05–2.47) | |||
| 12-D (TAG) | 25 (6.2) | 0.50 (0.70) | 5.3/6.3 | 1.10 (0.59–2.07) | |||
| Block free | |||||||
| F-A (GTCGAA) | 110 (25.5) | 0.42 (0.69) | <0.0001 | 30.5/20.6 | Reference | 0.022 | |
| F-B (GTCGAG) | 31 (7.2) | 0.48 (0.73) | 6.6/7.6 | 1.86 (0.98–3.54) | |||
| F-C (GTTAAG) | 30 (6.7) | 0.83 (0.73) | 5.5/8.3 | 2.35 (1.18–4.66) | |||
| F-D (GTTAGG) | 104 (24.3) | 0.57 (0.67) | 23.2/25.9 | 1.70 (1.09–2.66) | |||
| F-E (AGTAGG) | 42 (9.9) | 0.93 (0.80) | 8.4/11.5 | 2.19 (1.24–3.88) | |||
| No. of adverse haplotypes | |||||||
| 0 | 110 (25.5) | 0.42 (0.69) | 0.001 | 30.5/20.6 | Reference | 0.0061 | |
| 1–2 | 73 (17.2) | 0.57 (0.74) | 9.5/10.1 | 1.87 (1.05–3.62) | |||
| 3–4 | 172 (40.3) | 0.70 (0.78) | 37.3/43.1 | 1.94 (1.18–2.65) | |||
| 5–6 | 55 (12.6) | 0.92 (0.81) | 11.9/13.7 | 2.28 (1.19–3.99) | |||
Loci chosen for block 9: rs2256608, rs10483814, rs4899246, rs963918, rs963917 and rs10136316; Block 10: rs2064827, rs2145157, rs12878344 and rs17105965; Block 11: rs2331779, rs2331780 and rs12432197; Block 12: rs10438159, rs12893578 and rs4902623; block free: rs2256608, rs2064827, rs12432197, rs12893578, rs4902623 and rs2180611.
Haplotypes with frequency <0.05 were excluded from the analysis.
Dichotomized at the median of B/C (0.48), analysis adjusted by age and sex.
Generated by 10 000 permutation test.
Finally, we explored the haplotype association between mutagen sensitivity and six tSNPs of RAD51L1 globally using a block-free approach. Consistent with the genotype analysis, the most common haplotype, F-A, was also the most favorable haplotype, containing all six favorable alleles (zero adverse alleles) and exhibiting the lowest B/C value (Table III). When we used haplotype F-A as the reference group, haplotype F-C (containing three adverse alleles), haplotype F-D (containing four adverse alleles) and haplotype F-E (containing all six adverse alleles) all showed statistically significant increased mutagen sensitivity. The most adverse haplotype, F-E, completely mismatched at every SNP with the most favorable haplotype, F-A (termed ‘yin-yang haplotypes’), and exhibited the highest B/C value (Table III).
Further analyses of the number of adverse haplotypes revealed a significant dose–response relationship between the mutagen sensitivity and the number of adverse haplotypes. Mutagen sensitivity B/C values for individuals carrying zero, one to two, three to four or five to six adverse haplotypes were 0.42, 0.57, 0.70 and 0.92, respectively (trend test, P = 0.001). Using individuals carrying zero adverse haplotype as the reference group, the risks of being mutagen sensitive (B/C values greater than the median) was 1.87 (95% CI, 1.05–3.62) for those carrying one to two, 1.94 (95% CI, 1.18–2.65) for those carrying three to four and 2.28 (95% CI, 1.19–3.99) for those carrying five to six adverse haplotypes.
Replication results.
We further tested the six RAD51L1 SNPs (rs2256608, rs2064827, rs4902623, rs12432197, rs12893578 and rs2180611) in 662 healthy controls.The confirmation analysis was adjusted for age and sex. Only RAD51L1 rs2180611 showed significant association with mutagen sensitivity (Table III). Compared with individuals with the wild-type homozygous phenotype, those with the variant-type homozygous and heterozygous exhibited significantly lower B/C values (P = 0.025).
Individuals wild-type homozygous for RAD51L1 rs2180611 exhibited significantly higher gamma-radiation-induced B/C than individuals with the variants homozygous or heterozygous (0.33 versus 0.31). None of the SNPs were significantly different between the mutagen-sensitive and non-sensitive subgroups when dichotomized at the median of B/C value of 0.3 (Table IV).
Table IV.
Association between strand break repair pathway RAD51L1 gene SNP genotype and mutagen sensitivity in the replication control group
| SNP ID | Genotype | Controls (N = 662) |
Comparisona |
||||
| n (%) | B/C mean (SD) | P | Non-sensitive/sensitive | OR (95% CI) | P | ||
| Risk effect | |||||||
| rs2256608 | AG/GG | 634 (0.96) | 0.32 (0.15) | 0.721 | 314/320 | 1 (reference) | |
| AA | 28 (0.04) | 0.33 (0.17) | 12/16 | 1.33 (0.62–2.88) | 0.464 | ||
| rs2064827 | TT/TG | 633 (0.96) | 0.32 (0.15) | 0.282 | 313/320 | 1 (reference) | |
| GG | 29 (0.04) | 0.35 (0.18) | 13/16 | 1.18 (0.56–2.50) | 0.668 | ||
| rs4902623 | AA/AG | 551 (0.83) | 0.32 (0.15) | 0.347 | 274/277 | 1 (reference) | |
| GG | 110 (0.17) | 0.33 (0.15) | 51/59 | 1.19 (0.78–1.79) | 0.418 | ||
| Protective effect | |||||||
| rs12432197 | TT | 163 (0.25) | 0.33 (0.19) | 0.209 | 83/80 | 1 (reference) | |
| TC/CC | 499 (0.75) | 0.32 (0.13) | 243/256 | 1.09 (0.76–1.56) | 0.636 | ||
| rs12893578 | AA | 223 (0.34) | 0.33 (0.17) | 0.206 | 107/116 | 1 (reference) | |
| AG/GG | 439 (0.66) | 0.31 (0.14) | 219/220 | 0.91 (0.66–1.26) | 0.568 | ||
| rs2180611 | GG | 243 (0.37) | 0.33 (0.17) | 116/127 | 1 (reference) | ||
| AG/AA | 419 (0.63) | 0.31 (0.13) | 0.025b | 210/209 | 0.88 (0.64–1.21) | 0.416 | |
Dichotomized at the median of B/C (0.3), analysis adjusted by age and sex.
Bold numbers indicate significant values at P <0.05.
Discussion
In our comprehensive pathway-based evaluation of associations of variants of BER and DSBR genes with mutagen sensitivity, we found that seven of 392 selected tSNPs showed a robust association with mutagen sensitivity, of which six were from RAD51L1 and the other was from EME2. In addition, a more consistent and stronger correlation was observed between the mutagen sensitivity phenotype and the combination of these polymorphisms. Specifically, when the seven SNPs were analyzed together, we observed a significant dose–response relationship between the number of adverse genotypes and the mutagen sensitivity. Furthermore, haplotype analysis revealed significant differences in the haplotype profile for RAD51L1. The RAD51L1 haplotypes F-A (zero adverse alleles) and F-E (all six adverse alleles) have complementary nucleotides at all six sites, make opposing haplotypes and exhibited the lowest (B/C, 0.42) and highest (B/C, 0.93) mutagen sensitivity, respectively. Analyses of the number of adverse haplotypes revealed similar dose–response trend between the mutagen sensitivity and the number of adverse haplotypes. The association of RAD51L1 rs2180611 with mutagen sensitivity was confirmed in the validation set which contain 662 healthy control subjects.
Genetic instability can be triggered by the failure to repair breaks in double-stranded DNA that can be damaged by exposure to ionizing radiation. However, until now, no study has examined the effects of genetic polymorphisms of DSBR pathway genes on the mutagen sensitivity phenotype in patients with glioma or other cancers. We reported previously that increased sensitivity to radiation is an independent risk factor for gliomas (3,4). In the present phenotype–genotype analysis in glioma patients, we observed a strong association between polymorphisms in the DSBR pathway gene RAD51L1 and mutagen sensitivity.
The involvement of RAD51L1 in radiation-induced DNA damage is biologically plausible. RAD51L1 (also known as Rad51B, OMIM 602948), is a member of the RAD51 paralogue family (RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3) and plays a crucial role in homologous recombination of the DSBR pathway (19–21). Human RAD51L1 is induced by both ionizing and ultraviolet radiation, and its overexpression causes a delay in the G1 phase of the cell cycle and cell apoptosis (22). RAD51L1 has been shown to interact with RAD51C (23–25), which further interacts with the other family members, such as RAD51, XRCC2 and XRCC3 (26,27). Furthermore, RAD51L1 has been shown to interact directly with p53 (28), suggesting that this protein plays a role in the cell cycle, early embryonic development and perhaps apoptosis. An interesting feature of RAD51L1 in cancer genetics is that the gene maps to the chromosome break point in some benign tumors that harbor balanced chromosome translocations involving 14q23–24. This gene was reported to form fusion products upon translocation with HMGA2 (high-mobility-group protein family, member 2; OMIM 600698) in uterine leiomyoma (29). Interesting, HMGA2 was recently proved to be directly involved in DSBR pathway (30,31). RAD51L1 is also involved in the frequently occurring t (6;14) (p21;q23→q24) in pulmonary chondroid hamartomas (32). In addition, a recent genome-wide association study identified RAD51L1 as a new susceptibility gene for breast cancer risk (33). It is conceivable that the genetic variants of RAD51L1 may modify the association between breast cancer and radiation because it is up-regulated in human lymphocytes at radiation doses as low as 25 cGy (34). Furthermore, a copy number variant on chromosome 14q24.1 that includes RAD51L1 has been observed repeatedly in pedigrees with Li-Fraumeni syndrome with p53 germ line mutations, suggesting a possible contribution of this locus to the spectrum of cancers (including glioma) observed in this hereditary syndrome (35). Together, these studies strongly support a significant role for RAD51L1 in not only signaling and repairing DSBs induced by radiation but also genetic instability and cancer.
Interestingly, the in silico analysis using the SNP Function Portal server (http://brainarray.mbni.med.umich.edu/Brainarray/Database/SearchSNP/snpfunc.aspx) (18) revealed that RAD51L1 rs2256608 (intron 9) may be a recombination hot spot. Recent studies have shown that recombination is a ubiquitous feature of the human genome, and hot spots are the main contributor of the block-like pattern of haplotypes. A SNP in the vicinity of recombination hot spots could vastly increase the diversity of RAD51L1, a gene that is already diverse by virtue of polymorphisms. In addition, the SNP Function Portal (18) also identified the other five significant tSNPs of RAD51L1, which are all located in 3′ UTR region (i.e. rs2064827, rs12432197, rs12893578, rs4902623 and rs2180611), as transcription factor binding sites. It is plausible that these 3′ UTR polymorphisms may affect RAD51L1 expression by affecting either its RNA half-life or influencing the ribosomal translation of its messenger RNA. Therefore, these 3′ UTR polymorphisms may thus reduce the level of RAD51L1 protein and impair the cells’ capacity to protect the integrity of the genome upon environmental insult. However, the in silico functional prediction for these SNPs needs to be tested in vivo or in vitro.
Despite the strong and consistent associations of the tSNPs in RAD51L1 with mutagen sensitivity and the in silico predicted functional consequences of these SNPs, we recognize several potential limitations with the study. The main one is that most of our findings did not reach a statistically significant level in the replication set. However, replication failure should not be surprising or be interpreted as necessarily refuting the initial findings because of the potential problems such as population stratification and genetic heterogeneity (36–38). Our study subjects are glioma cases, whereas the replication study subjects are healthy controls. Although the dose of gamma-radiation used in the mutagen sensitivity assay in the two studies were almost the same (1.5 versus 1.25 Gy), their mean B/C were totally different (0.48 in our data set versus 0.3 in the replication set). When comparing the characteristics of the two groups, we observed large discrepancies in age (median 45.5 years in our cases versus 62.9 in the replication set). Therefore, additional information from other lines of evidence, such as resequencing and fine mapping of the interested RAD51L1 haplotype blocks, especially the blocks 11 and 12, the 3′ UTR region, followed by functional characterization studies to assess whether RAD51L1 is a true modifier gene and to identify the causal variations, will be more useful for validating and illuminating the functional relevance of genes identified in our study.
Another potential limitation is the study design. Tumor burden is a potentially important confounding factor in the measurement of DRC; the high metabolic rate and excessive endogenously generated oxidative stress in tumors might either suppress or enhance the DRC of lymphocytes. Therefore, a cohort study that measures repair prior to the development of cancer would be an ideal study design to make a definitive conclusion about the relationship between DRC and cancer risk (39).
In summary, our study revealed a strong correlation between polymorphisms and haplotypes of the RAD51L1 gene and mutagen-sensitivity phenotype in glioma patients. Our results support our earlier preliminary findings, which indicate that the sensitivity to gamma-radiation and the subsequent inability to repair radiation-induced DSBs as measured by chromatid breaks may increase the risk for brain tumorigenesis. Identification of causal variations in the RAD51L1 gene that account for the mutagen-sensitivity phenotype will help us to clarify the molecular mechanisms underlying mutagen sensitivity.
Supplementary material
Supplementary Tables 1, 2 and 3 can be found at http://carcin.oxfordjournals.org/
Funding
This work was supported by National Institutes of Health grants (5R01CA119215, 5R01CA070917). Additional support was obtained from the American Brain Tumor Association and the National Brain Tumor Society. The Wellcome Trust provided principal funding for the study. The replication study was supported by National Cancer Institute (CA98897, CA74880).
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- B/C
breaks per cell
- BER
base excision repair
- CI
confidence interval
- DRC
DNA repair capacity
- DSB
double-strand breaks
- DSBR
double-strand break repair
- FPRP
false-positive report probability
- LD
linkage disequilibrium
- OR
odds ratios
- SD
standard deviation
- SNP
single-nucleotide polymorphism
- tSNP
tag single-nucleotide polymorphism
- UTR
untranslated region
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Hsu TC, et al. Sensitivity to genotoxic effects of bleomycin in humans: possible relationship to environmental carcinogenesis. Int. J. Cancer. 1989;43:403–409. doi: 10.1002/ijc.2910430310. [DOI] [PubMed] [Google Scholar]
- 3.Bondy ML, et al. Mutagen sensitivity and risk of gliomas: a case-control analysis. Cancer Res. 1996;56:1484–1486. [PubMed] [Google Scholar]
- 4.Bondy ML, et al. Gamma-radiation sensitivity and risk of glioma. J. Natl Cancer Inst. 2001;93:1553–1557. doi: 10.1093/jnci/93.20.1553. [DOI] [PubMed] [Google Scholar]
- 5.Shete S, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, et al. Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol. Biomarkers Prev. 2009;18:204–214. doi: 10.1158/1055-9965.EPI-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ron E, et al. Tumors of the brain and nervous system after radiotherapy in childhood. N. Engl. J. Med. 1988;319:1033–1039. doi: 10.1056/NEJM198810203191601. [DOI] [PubMed] [Google Scholar]
- 8.Little MP, et al. Risks of brain tumour following treatment for cancer in childhood: modification by genetic factors, radiotherapy and chemotherapy. Int. J. Cancer. 1998;78:269–275. doi: 10.1002/(SICI)1097-0215(19981029)78:3<269::AID-IJC1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 9.Wood RD, et al. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 10.Akaike H. A new look at the statistical model identification. IEEE Trans. Automatic Control. 1974;19:716–723. [Google Scholar]
- 11.Lewontin RC. On measures of gametic disequilibrium. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 13.Schaid DJ, et al. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Human Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelman A, et al. Bayesian Data Analysis. 2nd edn. 2003. Chapman & Hall/CRC, p. 582. [Google Scholar]
- 15.Wacholder S, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, et al. Projecting individualized probabilities of developing bladder cancer in white individuals. J. Clin. Oncol. 2007;25:4974–4981. doi: 10.1200/JCO.2007.10.7557. [DOI] [PubMed] [Google Scholar]
- 17.Wu X, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P, et al. SNP Function Portal: a web database for exploring the function implication of SNP alleles. Bioinformatics. 2006;22:e523–e529. doi: 10.1093/bioinformatics/btl241. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama H, et al. Holliday junction binding activity of the human Rad51B protein. J. Biol. Chem. 2003;278:2767–2772. doi: 10.1074/jbc.M210899200. [DOI] [PubMed] [Google Scholar]
- 20.Thacker J. A surfeit of RAD51-like genes? Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- 21.Schild D, et al. Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- 22.Havre PA, et al. HsRec2/Rad51L1, a protein influencing cell cycle progression, has protein kinase activity. Exp. Cell Res. 2000;254:33–44. doi: 10.1006/excr.1999.4725. [DOI] [PubMed] [Google Scholar]
- 23.Miller KA, et al. RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J. Biol. Chem. 2002;277:8406–8411. doi: 10.1074/jbc.M108306200. [DOI] [PubMed] [Google Scholar]
- 24.Sigurdsson S, et al. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain S, et al. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum. Mol. Genet. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 26.Kurumizaka H, et al. Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proc. Natl Acad. Sci. USA. 2001;98:5538–5543. doi: 10.1073/pnas.091603098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu N, et al. Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res. 2002;30:1009–1015. doi: 10.1093/nar/30.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu Z, et al. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53(-/-) background. Mol. Cell. Biol. 1999;19:8686–8693. doi: 10.1128/mcb.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heim S, et al. A specific translocation, t(12;14)(q14-15;q23-24), characterizes a subgroup of uterine leiomyomas. Cancer Genet. Cytogenet. 1988;32:13–17. doi: 10.1016/0165-4608(88)90305-6. [DOI] [PubMed] [Google Scholar]
- 30.Li AY, et al. Suppression of nonhomologous end joining repair by overexpression of HMGA2. Cancer Res. 2009;69:5699–5706. doi: 10.1158/0008-5472.CAN-08-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cleynen I, et al. The HMGA proteins: a myriad of functions (Review) Int. J. Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 32.Blank C, et al. Intragenic breakpoint within RAD51L1 in a t(6;14)(p21.3;q24) of a pulmonary chondroid hamartoma. Cytogenet. Cell Genet. 2001;95:17–19. doi: 10.1159/000057011. [DOI] [PubMed] [Google Scholar]
- 33.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fachin AL, et al. Gene expression profiles in human lymphocytes irradiated in vitro with low doses of gamma rays. Radiat. Res. 2007;168:650–665. doi: 10.1667/RR0487.1. [DOI] [PubMed] [Google Scholar]
- 35.Shlien A, et al. Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc. Natl Acad. Sci. USA. 2008;105:11264–11269. doi: 10.1073/pnas.0802970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan TM, et al. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007;297:1551–1561. doi: 10.1001/jama.297.14.1551. [DOI] [PubMed] [Google Scholar]
- 37.Gorroochurn P, et al. Non-replication of association studies: "pseudo-failures" to replicate? Genet. Med. 2007;9:325–331. doi: 10.1097/gim.0b013e3180676d79. [DOI] [PubMed] [Google Scholar]
- 38.Liu YJ, et al. Is replication the gold standard for validating genome-wide association findings? PLoS One. 2008;3:e4037. doi: 10.1371/journal.pone.0004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berwick M, et al. Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J. Natl Cancer Inst. 2000;92:874–97. doi: 10.1093/jnci/92.11.874. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.