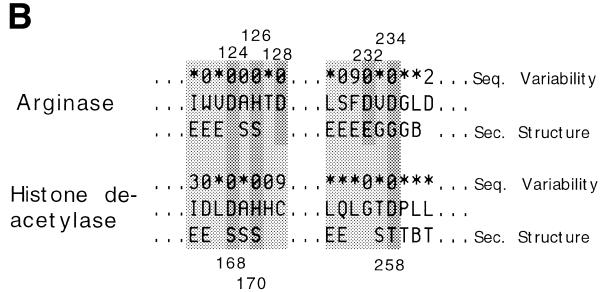
Figure 1.
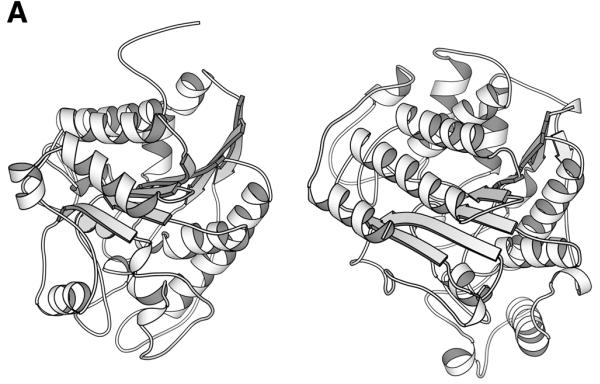
Unification of the histone deacetylase and arginase families. Reuse and adaptation of existing structural frameworks for new cellular functions is widespread in protein evolution. Histone deacetylase and arginase are unified at the functional family level of the classification despite very little overall sequence similarity. The supporting evidence comes from structural and functional similarity. (A) Structure comparison of arginase (left, 1rlaA) (10) and histone deacetylase (right, 1c3pA) (11) yields a high Z-score of 12. Superimposition by Dali, drawing by Molscript (12). (B) Joint structural, evolutionary and functional information for two segments around the active site. Structurally aligned positions are shaded. Arginase has a binuclear metal centre where residues D124, H126 and D234 bind one and residues H101, H128 and H232 the other manganese ion. The former site is structurally equivalent to the zinc binding site of histone deacetylase made up of residues D168, H170 and D258. Sequence variability from multiply-aligned sequence neighbours in HSSP (asterisk, values 10 or larger; 0, invariant) is shown above and the secondary structure summary from DSSP (E,B, beta-sheet, S bend; T,G, hydrogen-bonded turns) is shown below the amino acid sequences.