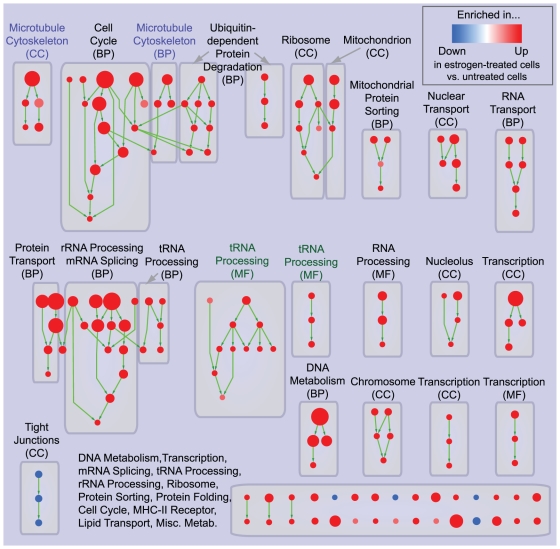
Figure 2. Hierarchical visualization of enrichment results for estrogen treatment of breast cancer cells.
Hierarchical organization of GO gene-set enrichment results for estrogen-treated compared to untreated breast cancer cells at 24 hours of culture. Nodes represent gene-sets and edges represent GO defined relations (Is-a, Part-of, Regulates). Gene-sets that did not pass the enrichment significance threshold are not shown. Nodes are colored according to enrichment results: red represents enrichment in estrogen-treated cells (i.e. up-regulation after estrogen treatment), whereas blue represents enrichment in untreated cells (i.e. down-regulation after estrogen treatment). Color intensity is proportional to enrichment significance. Since conservative thresholds were used to select gene-sets, most of the node colors are intense (corresponding to highly significant gene-sets). Subnetworks (i.e. clusters) are annotated according to the corresponding function. The acronym in brackets represents the specific GO ontology the gene-sets belong to: Molecular Function (MF), Cellular Component (CC), Biological Process (BP). Microtubule cytoskeleton (purple labels) and tRNA processing (green labels) were highlighted to show absence of connections between related gene-sets.