Abstract
Conclusions
The microarray analysis identified 39 genes up- or down-regulated by dexamethasone in the cultured tissue of mice cochlea. Of the eight genes most highly affected, several are suggested to have protective effects in the traumatized inner ear [Fkbp5, Glucocorticoid-induced leucine zipper (Gilz), glutathione peroxidase 3] and for others, a plausible mechanism of action can be offered (Claudin10, glutamate-ammonia ligase). The present data may support the use of dexamethasone to treat acute sensorineural hearing loss. It is warrantable to test these results in the in vivo cochlea.
Objectives
To identify genes of which expression are markedly up- or down- regulated by dexamethasone in the cochlear tissue.
Method
Murine cochlear tissue were cultured with or without dexamethasone for 48h in vitro. The gene expression profile were compared between the dexamethasone-treated and untreated cochlear tissue using a microarray that covers 33,696 transcripts (24,878 genes) of mice and quantitative realtime RT-PCR.
Results
The microarray analysis identified 39 genes that are up- or down-regulated by more than two-fold in the presence of dexamethasone in the cochlear culture. Genes up- or down-regulated by at least threefold include Fkbp5, Gilz, glutathione peroxidase 3, claudin 10, glutamate-ammonia ligase, proteoglycan1, integrin beta-like1 and alpha subunit of glycoprotein hormone.
Keywords: glucocorticoid, gene expressions, sensorineural hearing loss
Introduction
Glucocorticoid is widely used as a therapy for acute sensorineural hearing loss (SNHL), a practice supported by animal studies showing an inner ear protective effect in ischemia[1], sound trauma[2] and following exposure to aminoglycoside[3], cisplatin[4] and salicylate[5]. It is not fully understood how this protective effect is mediated, although glucocorticoid acts through the intracellular glucocorticoid receptor, a member of the nuclear receptor subfamily, to regulate gene transcription by DNA-binding dependent and independent pathways. The DNA-binding dependent pathway involves direct binding of the glucocorticoid receptor to glucocorticoid response elements (GRE) at regulatory regions of target genes; the DNA-binding independent pathway, in contrast, involves genes regulation through binding of glucocorticoid receptor to transcription factors and coactivators[6].
The explanted murine cochlea can be used to study gene expression in a variety of experimental conditions. During mouse development, the cochlear duct starts to differentiate from the otocyst at E12 from the dorsal walls of cochlear neuroepithelia. On E15, sensory hair cells and supporting cells have begun to differentiate[7], with ultrastructural maturation of hair cell stereocilia continuing until the second postnatal week. In earlier work we examined expression of several cochlear-specific deafness-associated genes including Tecta, Coch, Eya4 and Strc in E15 explants. The cochlear structures such as sensory epithelium and the expression of these genes were maintained in this explant especially within 48 h post dissection[8].
To determine the effect of glucocorticoid on the inner ear, we studied gene expression in murine cochlear explants treated with dexamethasone and compared these data to untreated controls using the Filgen Mouse 32k microarray that covers 24,878 murine genes.
Methods
Tissue dissection and culture of the mouse cochlea
Timed-pregnant female BALB/C mice were killed using ketamine (150mg/kg, i.p.) and cervical dislocation at E15 (vaginal plug, E0) where upon fetal cochleae were promptly dissected using a binocular microscope. The explanted cochlea included the cochlear duct and sensory epithelium, non-neuronal mesenchymal tissues, lateral wall and spiral neurons. All experimental protocols were in compliance with the guidelines of Okayama University’s Committee on Use and Care of Animals and Declaration of Helsinki.
Explanted cochleae were placed in 250 µℓ of a serum-free medium that contained Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 5% KnockOut Serum Replacement (Invitrogen, Carlsbad, CA, U.S.A) and penicillin G (0.06mg/ml), with or without dexamethasone (500ng/ µℓ). The medium was maintained at 37°C in 5% CO2 and changed daily. Cochleae were maintained in vitro for 48 h before extracting RNA. Usually, 8–12 cochleae from a single litter were processed in each sample. All experiments were performed in duplicate.
RNA extraction and probe labeling
Total RNA was purified from explanted cochlear tissue using an RNA easy column (Qiagen, Hilden, Germany) and treated with DNaseI. Cy-3 labeled antisense RNA probe was synthesized using RNA Transcript SureLABEL Core Kit ™ (Takara, Otsu, Japan). Two µgms total RNA was annealed with oligo dT primer at 70° for 6 m and subjected to reverse transcription using MLV reverse transcriptase at 42° for 1h. The RNA strand was digested by RNAseH and 2nd strand DNA was synthesized by E.coli DNA polymerase and E coli DNA ligase at 16° for 1 h. cDNA was purified by phenol:chloroform:isoamylalcohol extraction and ethanol precipitation. In vitro transcription and cRNA labeling was performed using Cy3-UTP and T7 RNA polymerase at 37° for 4 h.
Hybridization to the microarray
Three µgms of the labeled cRNA was diluted in 0.4M Tris-acetate (pH 8.2) and 0.3 M potassium acetate, heated at 94° for 10 m and cooled on ice. The probe solution was added to 10 µ g of Cot-I DNA (Invitrogen) and 10 µ g of yeast tRNA (Invitrogen) and ethanol-precipitated. The probe was diluted in 80 µℓ of the hybridization buffer (5 × SSC, 0.1%SDS, 10% formamide) and then incubated at 70° for 2 m, cooled to room temperature and applied on to the Filgen Mouse 32K array, that is a oligonucleotide microarray covering mouse 33,696 transcripts(24878 genes)(Filgen, Nagoya, Japan). After hybridization at 42° for 16–20 h, the array was washed in 0.1% SSC with 0.1% SDS at 25° for 1 m (× 4) and then dried by centrifugation at 25° and 200 × g for 5 m.
Microarray data scanning and analysis
The luminescence of the hybridized microarray was scanned using a GenePix 4000B scanner (Axon Instruments, Sunnyvale, CA, U.S.A.). The spot signals on the acquired TIFF image were quantitated with Array-pro Analyzer Software v4.5 (Media cybernetics, Bethesda, MD, U.S.A.) and defined as raw intensity. Net intensity was defined as [raw intensity – background value]. Normalized intensity of each signal spot was calculated by global normalization (normalization of net intensity to the mean net intensity of all signal spots). All experiments including probe labeling, hybridization, data scanning and analysis were performed in duplicate.
Quantitative real-time RT-PCR
Quantitative real-time RT-PCR was performed on genes up- or down-regulated at least three fold. Reverse transcription was performed with cochlear explant total RNA samples using random primers and MultiScribe™ reverse transcriptase from the High Capacity cDNA Archive kit (Applied Biosystems, Foster City. CA, U.S.A.), as described by the manufacturer. Specific PCR primers and the TaqMan probe for each gene were selected from pre-designed TaqMan Gene Expression Assay system (Applied Biosystems,) . The PCR reaction mix included gene-specific primers (900nM), 6-FAM™ dye-labeled TaqMan probe (250nM) and TaqMan Universal PCR Master Mix containing AmpliTaq Gold DNA polymerase. Hot-start two-step PCR consisting of preheat at 95° for 10 m, denature at 95° for 15 s, and annealing and extension at 60° for 1 m was performed using a 7500 Fast Real-time PCR systems (Applied Biosystems).
Relative abundance of the specific mRNAs in the cochlear explants was calculated from the difference of Ct value (cycling threshold, the minimum thermocycling number at which the fluorescence of 6-FAM™ released from the TaqMan probe became detectable). Data were normalized to the expression levels of internal controls, such as 18S rRNA.
Results
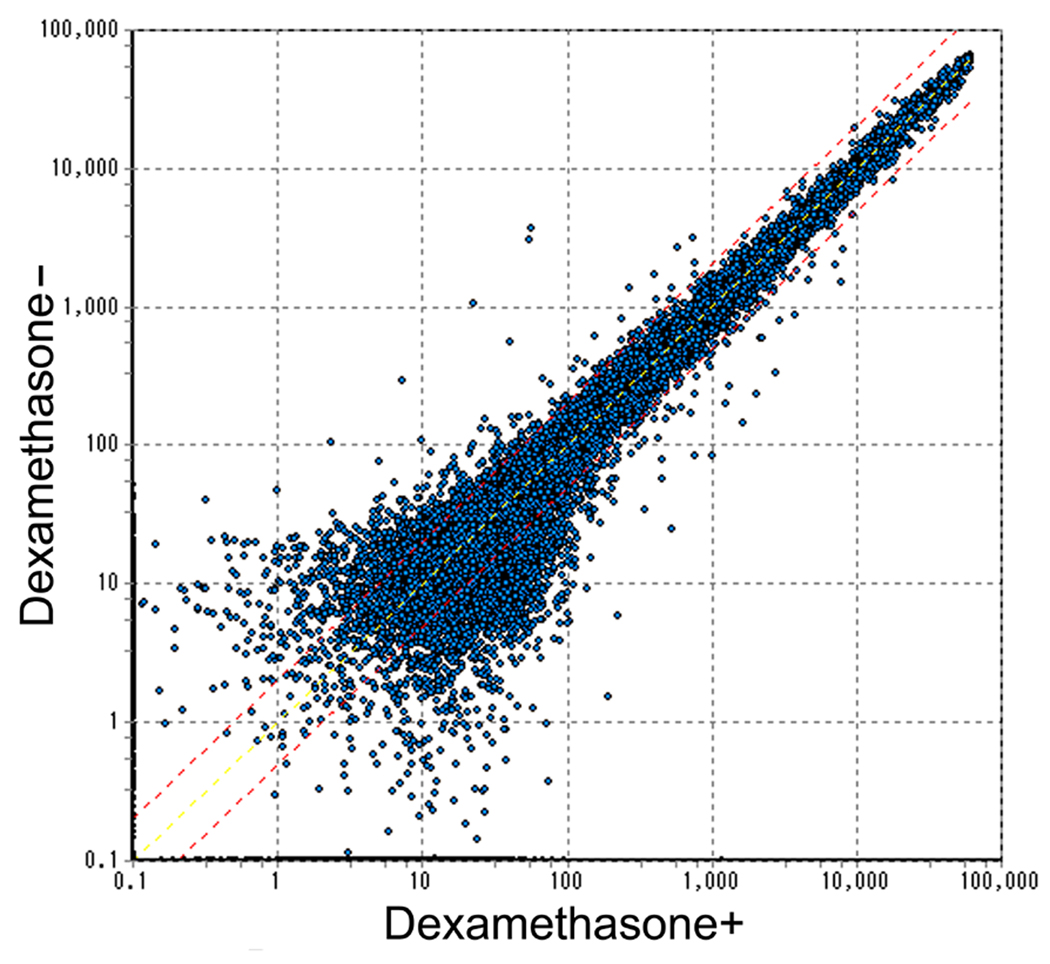
The overall correlation-of-expression of 33,696 transcripts (24,878 genes) between explanted cochlea with or without dexamethasone was extremely high (r=0.9928; Fig 1), validating the experimental conditions, however 39 genes were differentially up-regulated or down-regulated by more than two-fold in the presence of dexamethasone(Table 1). At least three-fold expression changes were noted for eight genes(Table 2). These results were confirmed by duplicate microarray experiments. For the genes of which expression were altered at least three fold, the results were also confirmed by quantitative real-time RT-PCR.
Fig. 1. Overall levels of gene expressions in the cochlear culture with or without dexamethasone.
The expression levels of the 33,696 transcripts (27,878 genes) were highly correlated between cochlear cultures with or without dexamethasone (r=0.9928).
Table 1.
Genes showing at least twofold change in expression in the presence of dexamethasone as compared to cochlear cultures without dexamethasone. The results are drawn from duplicated microarray experiments.
| Gene(RefSeq ID) | Dex(+) versus dex(−) ratio of normalized intensity. first trial |
Dex(+) versus dex(−) ratio of normalized intensity. second trial |
|---|---|---|
| cell division cycle associated 1(NM_023284) | 0.45 | 0.46 |
| caveolin, caveolae protein 1(NM_007616) | 2.24 | 3.86 |
| calcium binding protein 39-like(NM_026908) | 2.01 | 2.02 |
| procollagen, type VIII, alpha 1(NM_007739) | 0.49 | 0.43 |
| Lipopolysaccharide-binding protein precursor (LBP). (−) | 3.05 | 2.55 |
| kinesin family member 23(NM_024245) | 0.49 | 0.44 |
| matrix metalloproteinase 9(NM_013599) | 0.29 | 0.45 |
| FK506 binding protein 5(NM_010220) | 6.44 | 5.13 |
| proteoglycan 1, secretory granule(NM_011157) | 3.17 | 3.07 |
| glutamate-ammonia ligase (glutamine synthase)(NM_008131) | 3.13 | 4.27 |
| integrin, beta-like 1(NM_145467) | 5.00 | 3.05 |
| glycoprotein hormones, alpha subunit(NM_009889) | 0.23 | 0.27 |
| RIKEN cDNA 1110018M03 gene(NM_026271) | 3.80 | 2.72 |
| Glucocorticoid-induced leucine zipper protein. (NM_010286) | 3.54 | 3.58 |
| metallothionein 1(NM_013602) | 5.34 | 2.12 |
| keratin complex 2, basic, gene 6a(NM_008476) | 2.40 | 2.66 |
| glutathione peroxidase 3(NM_008161) | 3.71 | 4.49 |
| procollagen, type VI, alpha 1(NM_009933) | 2.03 | 3.40 |
| period homolog 1 (NM_011065) | 2.57 | 3.35 |
| P lysozyme structural(NM_013590) | 0.49 | 0.42 |
| Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal(−) | 3.35 | 2.06 |
| transforming, acidic coiled-coil containing protein 3(NM_011524) | 0.46 | 0.42 |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1(NM_016888) | 2.23 | 2.28 |
| tissue inhibitor of metalloproteinase 4(NM_080639) | 2.26 | 2.47 |
| carboxypeptidase Z(NM_153107) | 0.31 | 0.46 |
| claudin 11(NM_008770) | 2.29 | 3.48 |
| myelin and lymphocyte protein, T-cell differentiation protein(NM_010762) | 2.14 | 2.40 |
| basic transcription element binding protein 1(NM_010638) | 2.22 | 3.03 |
| cytoskeleton associated protein 2(NM_001004140) | 0.45 | 0.37 |
| S100 calcium binding protein A10 (calpactin)(NM_009112) | 2.08 | 3.22 |
| apolipoprotein D(NM_007470) | 2.16 | 2.38 |
| osteomodulin(NM_012050) | 2.44 | 2.14 |
| claudin 10(NM_021386) | 11.48 | 6.70 |
| LIM and senescent cell antigen like domains 2(NM_144862) | 2.80 | 3.03 |
| UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1(−) | 2.02 | 2.17 |
| protein phosphatase 1, regulatory (inhibitor) subunit 1B(NM_144828) | 2.68 | 2.17 |
| keratin complex 2, basic, gene 6a(NM_008476) | 2.19 | 2.05 |
| procollagen, type VI, alpha 2(NM_146007) | 2.15 | 2.39 |
| lipocalin 2(NM_008491) | 3.26 | 2.66 |
Table 2.
Genes up- or down-regulated by at least threefold in the presence of dexamethasone as compared to in vitro cochlear tissue without dexamethasone. The results from duplicated microarray experiments and quantitative real-time RT-PCR are shown.
| Gene(RefSeq ID) | Dex(+) versus dex(−) ratio of normalized intensity. first trial |
Dex(+) versus dex(−) ratio of normalized intensity. second trial |
Dex(+) versus dex(−) ratio of expression by quantitative realtime RT-PCR |
|---|---|---|---|
| FK506 binding protein 5 (NM_010220) | 6.44 | 5.13 | 6.19 |
| Glucocorticoid-induced leucine zipper protein. (NM_010286) | 3.54 | 3.58 | 3.64 |
| glutathione peroxidase 3(NM_008161) | 3.71 | 4.49 | 2.35 |
| claudin 10(NM_021386) | 11.50 | 6.70 | 5.64 |
| glutamate-ammonia ligase (MN_008131 (gluthamine synthetase) | 3.13 | 4.27 | 3.64 |
| proteoglycan1, secretory granule (NM_011157) | 3.17 | 3.07 | 2.82 |
| integrin beta-like1(NM_145467) | 5.00 | 3.05 | 3.93 |
| glycoprotein hormones, alpha subunit (NM_009889) | 0.23 | 0.27 | 0.22 |
Discussion
In this study, we looked at the effect of glucocorticoid on cochlear tissue and found that expression levels of 39 genes were altered by more than two-fold in dexamethasone-treated cochlear explants. At least three-fold expression changes were noted for eight genes. These genes are Fkbp5, Glucocorticoid-indeced leucine zipper(Gilz), glutathione peroxidase 3, claudin 10, glutamate-ammonia ligase, proteoglycan1, integrin beta-like1 and alpha subunit of glycoprotein hormone and interestingly, the biological function of some of these genes can be discussed in relation to hearing.
FK-binding protein 5 (FKBP5) is a binding partner that forms a tight complex with FK-506(Tacrolimus), a potent immunosuppressant[9]. It is reported that glucocorticoid administration upregulates FKBP5 in peripheral blood mononuclear cells[10] and that tacrolimus can cause SNHL in liver transplant recipients[11]. It is possible glucocorticoid blocks tacrolimus ototoxicity by upregulating FKBP5.
Glucocorticoid-induced leucine zipper (GILZ) is rapidly induced by dexamethasone in cultured cells[12]. This 137 amino acid protein inhibits Nuclear factor (NF) kappa beta by binding to its C-terminal PER domain[13]. If sudden SNHL activates cellular stress pathways involving NF-kappa beta[14], which is possible since NF-kappa beta translocates into the nucleus following noise-induced hearing loss in the cochlea[15], dexamethasone could exert a protective effect in the inner ear by activation of GILZ and inhibition of NF-kappa beta.
Glutathione peroxidase 3 catalyzes oxidation of glutathione and protects cells from oxidative damage. Systemic dexamethasone administration facilitates glutathione biosynthesis in murine spiral ganglion cells, which in turn attenuates the threshold shift and hair cell loss in noise-exposed guinea pigs[16].
Claudin 10 forms tight junctions in the organ of Corti, stria vascularis and Reissner’s membrane, and plays a critical role in maintaining the integrity of the scala media to facilitate the generation of the endocochlear potential[17, 18].
Glutamate-ammonia ligase (glutamine synthetase) catalyzes the conversion of glutamate and ammonia to glutamine, a cycle that is involved in replenishing the synaptic neurotransmitter glutamate. It is possible that glutamine synthetase functions to limit the perilymphatic glutamate concentration in the cochlea[19] .
Proteoglycan 1 is an extracellular matrix protein that was cloned from a human leukemia cells line, HL-60 and integrin beta-like 1 is a presumptive member of the integrin family of proteins. Integrins are extracellular matrix receptors and activate signal-transduction pathways that regulate actin cytoskeletal dynamics and cell motility. The alpha subunit of glycoprotein hormones is the common subunit of several dimeric proteins including chorionic gonadotropin (CG), luteinizing hormone (LH), follicle stimulating hormone (FSH) and thyroid stimulating hormone (TSH). It is difficult to speculate at this time how proteoglycan1, integrin beta-like 1 and the alpha subunits of glycoprotein hormones, contribute to the biological function of cochlea and hearing.
The limitation of this study is that we focused on developing cochlear explants. Glucocorticoid receptor acts as one of the transcritotional factors that regulate cochlear development[20]. The changes in the gene expressions observed in this study may be at least in part those associated with the developmental process of cochlea. However, in view of this limitation the data are quite remarkable. Of the eight genes most highly affected in this experimental situation, several are suggested to have protective effects in the traumatized inner ear (Fkbp5, GilZ, glutathione peroxidase 3) and for others, a plausible mechanism of action can be offered (Claudin10, glutamate-ammonia ligase). These data suggest that the use of dexamethasone in acute SNHL may be justified. It is warrantable to investigate dexamethasone-dependent expressions and biological function of these genes in the mature cochleae.
Acknowledgements
This work was supported by a grant from Ministry of Education, Culture Sports, Science and Technology of Japan (19591969) and in part by the NIH (NIDCD grant DC002842 (RJHS). RJHS is the Sterba Hearing Research Professor, University of Iowa College of Medicine.
Footnotes
There is no conflict of interest on this manuscript, including financial, consultant, institutional and other relations.
References
- 1.Morawski KTF, Bohorquez J, Niemczyk K. Preventing hearing damage using topical dexamethasone during reversible cochlear ischemia: an animal model. Otol. Neurotol. 2009;30:851–857. doi: 10.1097/MAO.0b013e3181b12296. [DOI] [PubMed] [Google Scholar]
- 2.Takemura K, Komeda M, Yagi M, Himeno C, Izumikawa M, Doi T, et al. Direct inner ear infusion of dexamethasone attenuates noise-induced trauma in guinea pig. Hear. Res. 2004;196:58–68. doi: 10.1016/j.heares.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Himeno C, Komeda M, Izumikawa M, Takemura K, Yagi M, Weiping Y, et al. Intra-cochlear administration of dexamethasone attenuates aminoglycoside ototoxicity in the guinea pig. Hear. Res. 2002;167:61–70. doi: 10.1016/s0378-5955(02)00345-3. [DOI] [PubMed] [Google Scholar]
- 4.Daldal A, Odabasi O, Serbetcioglu B. The protective effect of intratympanic dexamethasone on cisplatin-induced ototoxicity in guinea pigs. Otolaryngol. - Head & Neck Surgery. 2007;137:747–752. doi: 10.1016/j.otohns.2007.05.068. [DOI] [PubMed] [Google Scholar]
- 5.Park SK, Choi D, Russell P, John EO, Jung TT. Protective effect of corticosteroid against the cytotoxicity of aminoglycoside otic drops on isolated cochlear outer hair cells. Laryngoscope. 2004;114:768–771. doi: 10.1097/00005537-200404000-00033. [DOI] [PubMed] [Google Scholar]
- 6.Muzikar KA, Nickols NG, Dervan PB. Repression of DNA-binding dependent glucocorticoid receptor-mediated gene expression. Proc Natl Acad Sci U S A. 2009;106:16598–16603. doi: 10.1073/pnas.0909192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sher AE. The embryonic and postnatal development of the inner ear of the mouse. Acta. Otolaryngol. Suppl. 1971;285:1–77. [PubMed] [Google Scholar]
- 8.Maeda Y, Fukushima K, Kakiuchi M, Orita Y, Nishizaki K, Smith RJ. RT-PCR analysis of Tecta, Coch, Eya4 and Strc in mouse cochlear explants. Neuroreport. 2005;16:361–365. doi: 10.1097/00001756-200503150-00011. [DOI] [PubMed] [Google Scholar]
- 9.Weiwad M, Edlich F, Kilka S, Erdmann F, Jarczowski F, Dorn M, et al. Comparative analysis of calcineurin inhibition by complexes of immunosuppressive drugs with human FK506 binding proteins. Biochem. 2006;45:15776–15784. doi: 10.1021/bi061616p. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Burckart GJ, Shah T, Pravica V, Hutchinson IV. A novel method for monitoring glucocorticoid-induced changes of the glucocorticoid receptor in kidney transplant recipients. Transplant Immunol. 2009;20:249–252. doi: 10.1016/j.trim.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Norman K, Bonatti H, Dickson RC, Aranda-Michel J. Sudden hearing loss associated with tacrolimus in a liver transplant recipient. Transplant International. 2006;19:601–603. doi: 10.1111/j.1432-2277.2006.00317.x. [DOI] [PubMed] [Google Scholar]
- 12.Ayroldi ERC. Glucocorticoid-induced leucine zipper (GILZ): a new important mediator of glucocorticoid action. FASEB J. 2009;23:3649–3658. doi: 10.1096/fj.09-134684. [DOI] [PubMed] [Google Scholar]
- 13.Di Marco B, Massetti M, Bruscoli S, Macchiarulo A, Di Virgilio R, Velardi E, et al. Glucocorticoid-induced leucine zipper (GILZ)/NF-kappaB interaction: role of GILZ homo-dimerization and C-terminal domain. Nucleic Acids Res. 2007;35:517–528. doi: 10.1093/nar/gkl1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merchant SN, Adams JC, Nadol JB., Jr Pathology and pathophysiology of idiopathic sudden sensorineural hearing loss. Otol. & Neurotol. 2005;26:151–160. doi: 10.1097/00129492-200503000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Masuda M, Nagashima R, Kanzaki S, Fujioka M, Ogita K, Ogawa K. Nuclear factor-kappa B nuclear translocation in the cochlea of mice following acoustic overstimulation. Brain Res. 2006;1068:237–247. doi: 10.1016/j.brainres.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Ohinata Y, Yamasoba T, Schacht J, Miller JM. Glutathione limits noise-induced hearing loss. Hear. Res. 2000;146:28–34. doi: 10.1016/s0378-5955(00)00096-4. [DOI] [PubMed] [Google Scholar]
- 17.Kitajiri SI, Furuse M, Morita K, Saishin-Kiuchi Y, Kido H, Ito J, et al. Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hear. Res. 2004;187:25–34. doi: 10.1016/s0378-5955(03)00338-1. [DOI] [PubMed] [Google Scholar]
- 18.Souter M, Forge A. Intercellular junctional maturation in the stria vascularis: possible association with onset and rise of endocochlear potential. Hear. Res. 1998;119:81–95. doi: 10.1016/s0378-5955(98)00042-2. [DOI] [PubMed] [Google Scholar]
- 19.Eybalin M, Norenberg MD, Renard N. Glutamine synthetase and glutamate metabolism in the guinea pig cochlea. Hear. Res. 1996;101:93–101. doi: 10.1016/s0378-5955(96)00136-0. [DOI] [PubMed] [Google Scholar]
- 20.Nagashima R, Sugiyama C, Yoneyama M, Ogita K. Transcriptional factors in the cochlea within the inner ear. J. Pharmacol. Sci. 2005;99:301–306. doi: 10.1254/jphs.cpj05004x. [DOI] [PubMed] [Google Scholar]