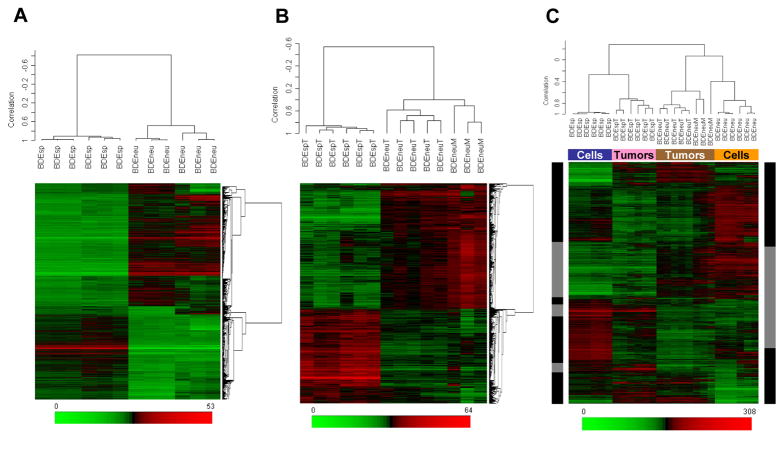
Figure 1.
Two-dimensional hierarchical clustering of samples and genes using Pearson’s (centered) correlation and average linkage, based on (A) 4,711 probe sets that were significantly different (q < 0.05) between BDEneu and BDEsp cultured cell lines, (B) 3,090 probe sets that were significantly different (q < 0.05) between BDEneu liver tumors and peritoneal metastases compared to non-metastatic BDEsp liver tumors, and (C) 4,711 probe sets that were significantly different between BDEneu and BDEsp cell lines used to cluster all of the samples: BDEsp cell line (blue box), BDEneu cell line (orange box), BDEsp tumors (pink box), and BDEneu tumors (brown box). The 1,497 common probe sets for cells in tumors are indicated by gray sidebars, whereas the discordant probe sets are indicated by black sidebars. Each of the colored rows in the individual heat maps show the relative expression for that specific gene in the separate specimen samples (columns). The relative gene expression levels are plotted according to the color scale at the bottom of each heat map, where red and green respectively indicate the relative extents of gene overexpression or underexpression compared to the median intensity across all samples.