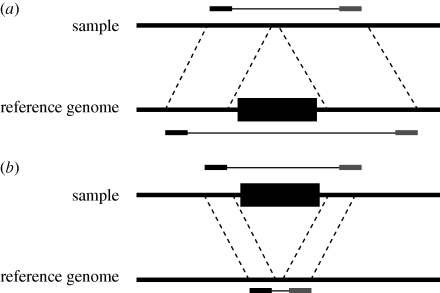
Figure 2.
Detecting insertions and deletions using paired-end mapping data. The ends of a DNA fragment from a sample individual are mapped to a reference genome. In both of these illustrations, a depiction of the DNA fragment appears above its location in the sample chromosome. The black and grey ends correspond to the unique sequenced ends of the fragment, and the expected length of the sequence is shown above the sample chromosome. Dashed lines indicate homologous regions in the two genomes, and the location of the black and grey ends below the reference chromosome corresponds to their mapped locations. (a) If the portion of the reference genome spanned by the fragment ends is larger than expected, then the sample genome probably contains a deletion relative to the reference. (b) If the length of the region spanned by the locations of the end sequences in the reference genome is smaller than expected, then an insertion is inferred to be present in the sample genome.