Figure 6.
PI4P Lipid Microenvironment within Replication Organelles Regulates both Enteroviral and Flaviviral RNA Replication
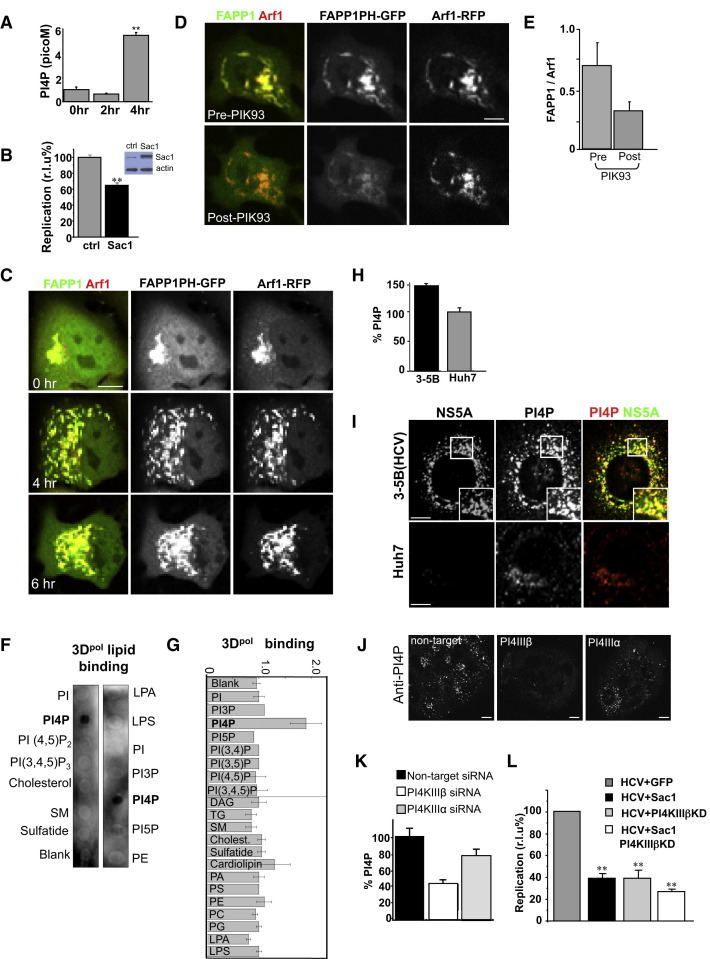
(A) Cellular PI4P lipid levels rise in CVB3 infection. Total cellular PI4P lipids were quantified over time. Error bars are SEM from duplicate samples (∗∗p < 0.01).
(B) Reduction in PI4P lipid levels inhibits enteroviral RNA replication. PV replicon assays in HeLa cells ectopically expressing Sac1 are shown. Inset: western blot showing increase in Sac1 levels after 16 hr of ectopic expression. Bar graph presents maximum replication values, normalized to control (GFP) plasmid ectopic expression. Error bars are SEM from eight replicates cells (∗∗p < 0.01).
(C) PI4P lipids localize to enteroviral replication organelles. Time-lapse confocal images of single HeLa cell infected with CVB3, coexpressing FAPP1PH-GFP/Arf1-RFP (see also Movie S3).
(D) PI4KIIIβ activity is responsible for PI4P lipids at enteroviral replication organelles. Time-lapse confocal image of single HeLa cell coexpressing FAPP1-PH-GFP/ARF1-RFP pre- and post-PIK93 treatment at 4 hr post-infection with CVB3.
(E) Quantification of FAPP1PH-GFP to Arf1-RFP fluorescence from (D) expressed as a ratio. Error bars are SEM from ten cells for each condition.
(F and G) PV RNA polymerase specifically and preferentially binds PI4P lipids. Purified recombinant 3Dpol enzyme was incubated with membrane strips that were previously spotted with different lipids. Antibodies detected 3Dpol binding. Two representative blots are shown. Binding was quantified and plotted in bar graph as a ratio over background non-lipid spotted membrane from three different experiments for each lipid type.
(H) Cellular PI4P lipid levels rise in HCV replicating cells. Quantification of PI4P with anti-PI4P primary antibodies of 3-5B(HCV) cells, normalized as % of PI4P lipid quantification within Huh7 cells. Error bars are SEM from ten cells for each cell type.
(I) PI4P lipids localize to HCV replication membranes. PI4P lipid and NS5A protein distribution in Huh7 and 3-5B(HCV) cells were determined by immunostaining with anti-PI4P and anti-NS5A antibodies.
(J and K) PI4KIIIβ is responsible for a significant fraction of PI4P lipids at HCV replication membranes. 3-5B(HCV) cells were treated with nontargeting, PI4KIIIβ, or PI4KIIIα siRNA. Cells were immunostained and quantified for PI4P lipids. Representative images of groups of siRNA-treated cells are shown (J). Quantification was done on 20 cells for each siRNA treatment condition.
(L) Reduction of PI4P lipids inhibits HCV replication. HCV replicon assays conducted with J6/JFH (p7-Rluc2A) replicons in Huh7 cells ectopically expressing Sac1, PI4KIIIβ-KD, or both plasmids are shown. Bar graph presents maximum replication value normalized to control (GFP) plasmid ectopic expression. Error bars are SEM from eight replicates of cells for each treatment condition (∗∗p < 0.01).
All fluorescence images were confocal images of optical slice thickness ∼1 μm. Scale bar, 10 μm.