Figure 4.
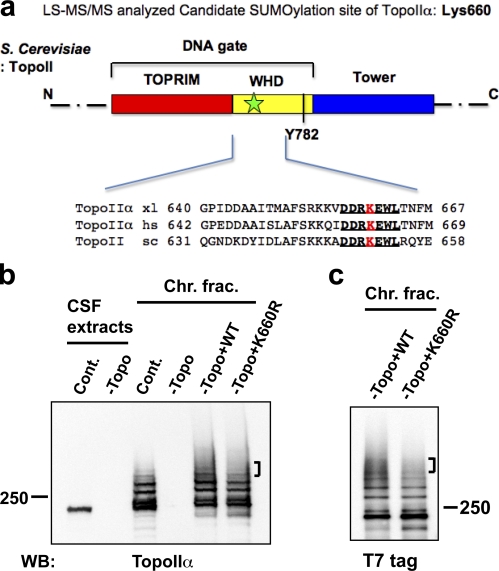
TopoIIα K660R, a candidate SUMOylation mutant, shows incomplete SUMOylation in XEE. (a) Schematic diagram of S. cerevisiae TopoIIα primary structure. Domains are denoted by color. This panel was modified from Schoeffler and Berger (2008), with permission from Cambridge University Press. TOPRIM, Topoisomerase-primase fold domain; WHD, Winged-helix domain; Tower, adjacent domain to WHD. The black bar indicates the catalytic tyrosine (Y782) for DNA cleavage in the WHD domain. Lys660 in X. laevis TopoIIα was designated as a potential SUMOylation site by mass spectrometric analysis. The approximate position of the candidate lysine is shown by a green star in the DNA gate domain of TopoIIα. The sequences near TopoIIα Lys660 from X. laevis (xl), Homo sapiens (hs), and S. cerevisiae (sc) are conserved (indicated with bold and underlined text). (b) XEE were immunodepleted using nonspecific IgG (Cont.) or an anti-TopoIIα antibody (−Topo). Efficiency of TopoIIα depletion was confirmed by comparison of mock-depleted (Cont.) to TopoIIα-depleted (−Topo) CSF extracts (left two lanes, labeled CSF extracts). WT nontagged TopoIIα (WT) or mutant TopoIIα, with substitution of arginine for Lys660 (K660R), was added to the TopoIIα-depleted extracts (−Topo). After 1 h incubation at 25°C, mitotic chromosomes were isolated and analyzed by anti-TopoIIα Western blotting. Analyzed chromosome samples were from mock-depleted (Cont.), TopoII-depleted (−Topo), and recombinant TopoIIα added-back extracts (−Topo+WT or –Topo+K660R). (c) Same examination as in b except that the recombinant TopoIIα proteins had a T7 tag at the N terminus. Both nontagged and T7-tagged K660R mutant showed subtle but reproducible reduction in higher shifted bands (indicated by brackets) of SUMOylation compared with WT. Positions of molecular mass standards (kD) are indicated on the sides of the gel blots.