Abstract
Serine protease HtrA1 belongs to a family of chymotrypsin-like proteases that were first identified in bacteria and later in mammalian systems. These proteases were identified as components of protein quality control in prokaryotic systems and as regulators of diverse signaling pathways in mammalian systems. In particular, HtrA1 is implicated in trophoblast cell migration and invasion, tumor progression, chemotherapy-induced cytotoxicity, osteoarthritis, age-related macular degeneration, and pathogenesis of Alzheimer’s disease. However, systematic analysis of its potential substrates in biological system is still lacking. Therefore, we performed a mixture-based oriented peptide library screening to identify putative substrates of HtrA1. We identified [AEGR]-[LAGR]-[IAMLR]-[TVIAL] as consensus residues for P1 to P4 sites. We identified several putative substrates of HtrA1 involved in the pathogenesis of various diseases. In this study, we report on the identification of tubulins as potential substrates of HtrA1, and validated tubulins as in vitro and intracellular substrates of HtrA1. These results provide initial insights into substrate identification and functional characterization of HtrA1 in pathogenesis of various diseases.
Keywords: Serine protease, HtrA1, Tubulins, Peptide library
The HtrA family of serine proteases was initially identified in E. coli by two phenotypes of null mutants that were unable to grow at elevated temperatures (HtrA for High temperature requirement) [Lipinska et al., 1988], or failed to digest misfolded protein in the periplasm (DegP) [Strauch and Beckwith, 1988]. Subsequently, homologues of HtrA/DegP have been described in a variety of species, including Gram-negative and -positive bacteria, plants and mammals. These proteins normally contain two conserved core domains, a chymotrypsin-like protease domain, and at least one C-terminal PDZ domain. In contrast to other protease-chaperone systems, HtrA represents the first well-known protein quality control factor that acts in an ATP-independent manner [Pallen and Wren, 1997; Spiess et al., 1999].
Until now, four human homologues of E. coli HtrA have been identified: HtrA1 (L56 or PRSS11) [Hu et al., 1998; Zumbrunn and Trueb, 1996], HtrA2/Omi [Faccio et al., 2000; Gray et al., 2000], HtrA3 (PRSP) [Nie et al., 2003] and HtrA4. All mammalian HtrA proteins, belonging to this family, share a highly conserved chymotrypsin-like serine protease domain and one PDZ domain at the C-terminus [Oka et al., 2004]. Otherwise, structure of N-terminal regions of mammalian HtrA1, 3 and 4 are distinct from that of HtrA2/Omi: mitochondrial HtrA2/Omi posses a transmembrane anchor, and a large section of the N-terminus is removed by processing, whereas the N-termini of HtrA1, 3 and 4 all contain predicted signal peptides as well as domains that are recognized as IGF binding and protease inhibitor domains [Clausen et al., 2002]. Although HtrA1 contains signal peptide, intracellular localization of HtrA1 has recently been reported [Clawson et al., 2008].
The HtrA family of serine protease appears to be involved in several important biological mechanisms in mammals, such as growth, apoptosis, arthritis, embryogenesis, neurodegenerative and neuromuscular disorder, and cancer. HtrA1 is the first sequenced member of the human HtrA protein family when it was identified as a gene expressed by normal fibroblasts but not by SV40 transformed counterparts [Zumbrunn and Trueb, 1996]. Subsequently, it was identified as a protein overexpressed in osteoarthritic cartilage [Hu et al., 1998]. HtrA1 has a widespread pattern of expression, and suggested to modulate several physiologic and pathophysiologic processes, such as TGF-β signaling [Oka et al., 2004], programmed cell death [Chien et al., 2006; Chien et al., 2004], cell proliferation [Baldi et al., 2002], trophoblast migration and invasion [Ajayi et al., 2008], osteoarthritis [Grau et al., 2006; Hu et al., 1998; Tsuchiya et al., 2005], tumor progression [Baldi et al., 2002; Chien et al., 2004], age-related macular degeneration [Yang et al., 2006], Alzheimer’s disease [Grau et al., 2005], and implantation [De Luca et al., 2003].
Extracellular matrix proteins, such as collagens, fibronectin, fibromodulin, and cytokines, such as TGFβ and BMPs, have been identified as potential substrates of HtrA1. To identify additional proteins that could act as potential substrates of HtrA1, we determined HtrA1 cleavage site motifs using a mixture-based oriented peptide library screening, and identified tubulins as potential substrates of HtrA1. These results provided potential insights into the functional role of HtrA1 in microtubule-related cell biology.
MATERIALS AND METHODS
Cell culture
SKOV3 and OV202 cells were grown as previously described [Chien et al., 2006]. Transfection was performed using Lipofectamine as previously described [Chien et al., 2006].
Generation of peptide libraries
The first random mixture of peptide library was generated as previously described [Turk and Cantley, 2004; Turk et al., 2001]. The second peptide library was generated as M-A-X-X-X-X-R-P-D-F-(K-biotin), where X represents totally degenerate amino acid residue, similar to procedure previously described [Turk and Cantley, 2004; Turk et al., 2001].
Primary peptide library screening and determination of primed side selectivity
Mixture-based oriented primary peptide library containing completely random octamer acetyl-XXXXXXXX-amide, where X indicates a degenerate position, was synthesized following previously described procedure [Turk and Cantley, 2004; Turk et al., 2001]. 2 mg/ml of this library was digested with 2 μg of purified recombinant HtrA1 for 3 hours at 37°C and processed as previously described before subjecting to amino terminal sequencing [Turk and Cantley, 2004; Turk et al., 2001]. Based on the molar proportion of each residue present within a given sequencing cycle, selectivity of amino acid for P1 to P4 relative to cleavage site is determined [Turk and Cantley, 2004; Turk et al., 2001].
Secondary peptide library screening and determination of unprimed side selectivity
Secondary peptide library containing partially degenerate M-A-X-X-X-X-R-P-D-F-K(biotin)-amide was synthesized following previously described procedure [Turk and Cantley, 2004; Turk et al., 2001]. This library was pre-purified with immobilized reversible-binding avidin column to remove peptides with no affinity to the column [Turk and Cantley, 2004; Turk et al., 2001]. 2 mg/ml of purified peptide library was digested with 2 μg of purified recombinant HtrA1 for 3 hours at 37°C and subjected to avidin affinity purification to remove un-reacted and C-terminal fragments of cleaved peptides as previously described [Turk and Cantley, 2004; Turk et al., 2001]. Unbound peptides were subjected to amino terminal sequencing to determine the selectivity of amino acids corresponding to P1-P4 of unprimed side as previously described [Turk and Cantley, 2004; Turk et al., 2001].
N-terminal sequencing
Sequencing was performed as previously described [Turk and Cantley, 2004; Turk et al., 2001].
In vitro digest
Purified tubulins (Cytoskeleton, Denver, CO) were incubated with varying concentrations of purified HtrA1 for 3 h at 37°C. Reaction products were resolved on SDS-PAGE and either stained with Coomassie blue or immunoblotted with specific tubulin antibodies. 2 μg of purified collagens (Rockland Immunochemicals, Inc., Gilbertsville, PA) were incubated with specified concentrations of purified HtrA1 for 3 h at 37°C. Reaction products were resolved on SDS-PAGE and stained with Coomassie blue.
Antibodies
Polyclonal antibodies raised against a polypeptide corresponding to amino acids 161-480 of HtrA1 were affinity purified as previously described [Chien et al., 2006]. Monoclonal antibodies against β-actin and acetylated and non-acetylated α-tubulin were purchased from Sigma-Aldrich.
Expression constructs
Protease active (WT) and protease inactive mutant (SA) HtrA1 construct in pcDNA3.1 plasmids were generated as previously described [Hu et al., 1998]. GFP-fusion constructs were generated by PCR cloning into pcDNA3.1/TOPO/GFP (Invitrogen).
Purification of recombinant HtrA1
Human recombinant HtrA proteins were purified from bacteria as previously described [Grau et al., 2006].
Transfection with siRNA
Scrambled control siRNA (5’-UCCUGCUGGAGCCUCAUGUTT-3’) and HtrA1 siRNA targeting the 3’ UTR (5’- CGGCCGAAGUUGCCUCUUUTT-3’) were purchased from Proligo LLC (Boulder, CO) and transfected at the final concentration of 0.25 μM in OptiMEM using Oligofectamine (Invitrogen). SKOV3 cells were plated at 20,000 cells/well in 24-well plate or at 100,000 cells/well in 6-well plate for transfection with siRNA. Effectiveness of RNAi was determined by immunoblot after 2 day of transfection. Using standard conditions provided by the manufacturer, we typically achieved >90% reduction in HtrA1 expression after 2 day of transfection.
Rescue of RNAi
For rescue of RNAi, cells were initially transfected with siRNA. Twenty four hours later, cells were transfected with plasmid carrying HtrA1 ORF. At the same time, control groups received mock transfections. Since RNAi is targeted toward the 3’ UTR of HtrA1 mRNA, the mRNA encoded from the plasmid containing HtrA1 ORF does not contain the 3’UTR and therefore is resistant to RNAi.
Immunostaining and fluorescence microscopy
Immunostaining was performed as described previously [Lingle and Salisbury, 2001]. Laser-scanning confocal microscopy was performed on a Zeiss LSM510 with krypton–argon and helium–neon lasers.
Analysis of tubulin immunofluorescence
Fluorescence intensity was analyzed by first converting red fluorescence signal from raw images to grayscale and analyzing the pixel intensity by KS400 Imaging System software (Zeiss). Total fluorescence intensity and surface area of at least 40 cells were measured. Average intensity of each cell was obtained by dividing total intensity with surface area. Data were represented as mean measurement ± s.e.m of 40 cells.
Flow cytometry
SKOV3 cells transfected with either control siRNA or HtrA1-targeting siRNA, or rescued with ectopic expression of HtrA1 were fixed, permeabilized, stained with antibody against α-tubulin, and analyzed by flow cytometry.
Immunoblotting
Cells were lysed in buffer (20 mM Tris-HCl at pH 7.6, 120 mM NaCl, 1 mM EDTA, 0.5% Nonidet P40, 1 mM dithiothreitol) supplemented with complete protease inhibitors (Roche). Whole cell lysates and cytosolic extracts were analyzed by western blot with anti-HtrA1 antibody (dilution 1:1,000), anti-β-actin antibody (1:5,000), anti-α-tubulin (1:1000), and anti acetylated-α-tubulin (1:1000).
Densitometric analysis
Immunoblots from three independent experiments were analyzed by Scion Image software (Frederick, MD). Arbitrary intensity values were normalized to β-actin loading controls and expressed as fold-change over DMSO-treated SKOV3 cells stably transfected with non-targeting shRNA (NT). Mean±sem are shown in bar graphs.
Statistical analysis
Significance of change in expression values (expressed as fold-change) for α-, β-, and γ-tubulins following HtrA1 knockdown was analyzed by Analysis of Varian (ANOVA) followed by Newman-Keuls Multiple Comparison Test with p<0.05 for significance. Significance of change in expression values for α-, β-, and γ-tubulins following Taxol and Nocodazole treatment was analyzed by ANOVA followed by Dunnett’s Multiple Comparison Test with p<0.05 for significance. Dunnett’s test allows comparison of all groups vs. control group (DMSO-treated SKOV3 cells stably transfected with non-targeting shRNA).
RESULTS
Identification of consensus P1’ to P4’ cleavage site motif using completely degenerate peptide library
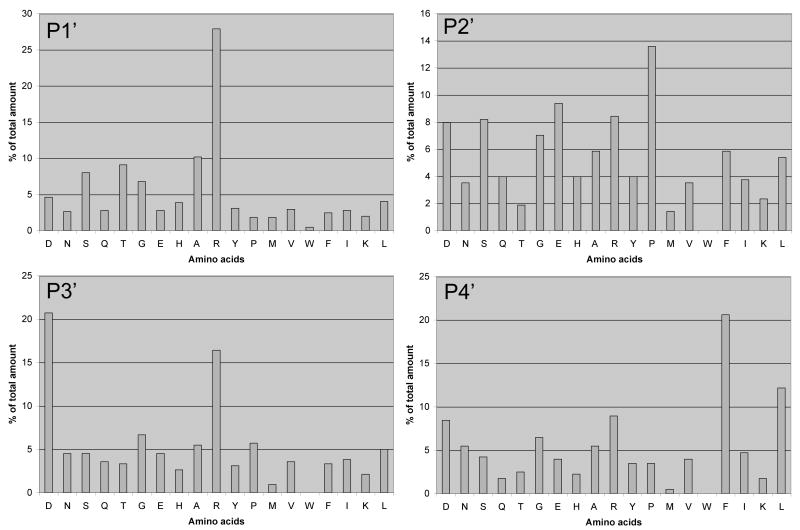
Amino-terminal acetylated peptide library was incubated with recombinant HtrA1 for 3 h, and P1’ to P4’ consensus residues of cleaved peptides were identified by N-terminal sequencing by Edman degradation. Uncleaved intact peptides and the N-terminal fragments of cleaved peptides remain blocked and do not contribute to the sequencing results. In contrast, newly generated N-termini of cleaved C-terminal fragments can be sequenced. The relative distributions of each amino acid present in a given cycle indicate the preference for such residues at a particular site. For example, the first sequencing cycle determines consensus (most) preferred residues for the P1’ position, the second cycle for the P2’ position, and so on. The results, shown in Figure 1, indicate R-P-D-F as the consensus motif occupying P1’ to P4’, respectively.
Fig. 1.
Identification of consensus HtrA1 cleavage motif for primed positions. 2 mg/ml of purified peptide library was digested with 2 μg of purified recombinant HtrA1 for 3 hours at 37°C and subjected to N-terminal sequencing. Since N-termini of all peptides in the library are blocked, only the cleaved peptides contribute to sequencing. Based on the molar proportion of each residue present within a given sequencing cycle, selectivity of amino acid for P1 to P4 relative to cleavage site is determined [Turk and Cantley, 2004; Turk et al., 2001].
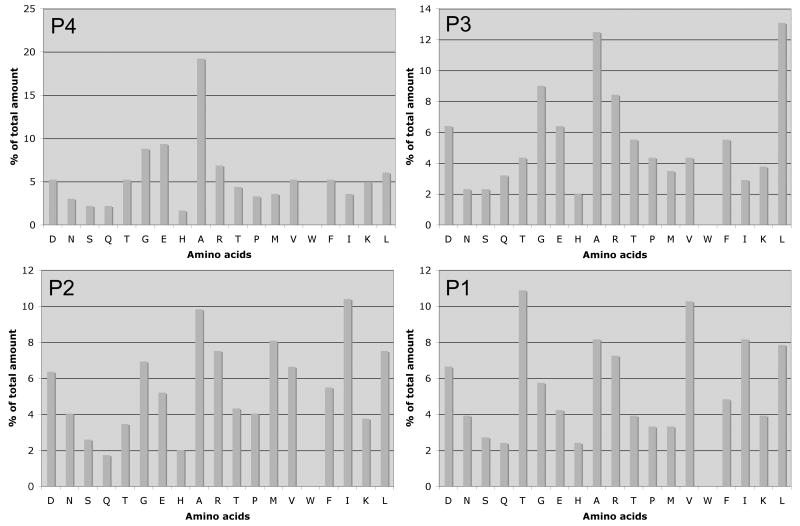
Identification of consensus P1 to P4 cleavage site motif using oriented second peptide library
Based on the consensus primed-side motifs, we generated the second library containing mixture of peptides with the following motif: MAXXXXRPDF(K-biotin), where X indicates a degenerate position, K-biotin is ε-(biotinamidohexanoyl)lysine, and the N terminal is unblocked. The fixed RPDF represents the consensus primed-side motif, whereas residues occupying unprimed (P1 to P4) positions are totally degenerate. This approach affords determination of consensus unprimed motif for HtrA1. Following digestion with HtrA1, unreacted (uncleaved) peptides and cleaved C terminal fragments are removed from the reaction by avidin column, and flow through, containing N-terminal fragments of cleaved peptides were subjected to N-terminal sequencing by Edman degradation. The results, shown in Figure 2, indicate [AEGR]-[LAGR]-[IAMLR]-[TVIAL] as the consensus unprimed motifs for HtrA1 substrates.
Fig. 2.
Identification of consensus HtrA1 cleavage motif for unprimed positions. 2 mg/ml of purified peptide library was digested with 2 μg of purified recombinant HtrA1 for 3 hours at 37°C and subjected to avidin affinity purification to remove un-reacted and C-terminal fragments of cleaved peptides as previously described [Turk and Cantley, 2004; Turk et al., 2001]. Unbound peptides were subjected to amino terminal sequencing to determine the selectivity of amino acids corresponding to P1-P4 of unprimed side as previously described [Turk and Cantley, 2004; Turk et al., 2001].
Determination of potential substrates of HtrA1
To determine the potential substrates containing the consensus unprimed motifs, [AEGR]-[LAGR]-[IAMLR]-[TVIAL] motif was searched using ProScan website, and potential substrates were identified (Table 1). Putative substrates identified through aforementioned approach are categorized into classes of proteins or their pathologic association to specific disease groups (Table 1).
Table 1.
Representative list of putative substrates identified from the peptide library
| Apoptosis Regulators | Cytoskeletal Proteins | Adhesion Proteins | MDR Pumps | Accessible/Total |
|---|---|---|---|---|
| Total Hits 106 | Total Hits 99 | Total Hits 47 | Total Hits 9 | Cut sites |
| APR3 (Q6UW56) | KRT10 (P13645) | CD44 (P16070) | MDR1 (P08183) | 11/13 |
| IAP1 (Q13489) | KRT12 (Q99456) | CEACAM19 (Q7Z692) | MDR3 (P21439) | 11/11 |
| IAP2 (Q13490) | KRT13 (P13646) | FAK1(Q05397) | ABCB5 (Q2M3G0) | 11/11 |
| IAP3 (P98170) | KRT14 (P02533) | FAK2(Q14289) | ABCB6 (Q9NP58) | 8/14 |
| LIVIN (Q96CA5) | KRT15 (P19012) | ICAM1 (P05362) | ABCB7 (O75027) | 10/11 |
| SIVA (O15304) | TUBA (alpha) | ICAM3 (P32942) | ABCB8 (Q9NUT2) | 9/14 |
| TAIP2 (Q8WYN3) | TUBB (beta) | ICAM4 (Q14773) | ABCB9 (Q9NP78) | 8/8 |
| BIRC1 (Q13075) | TUBG (gamma) | ICAM5 (Q9UMFO) | ABCB10 (Q9NRK6) | 9/14 |
| BIRC5 (O15392) | TPPP1 (O94811) | ARHGAP10 (A1A4S6) | ABCB11 (O95342) | 16/19 |
| BIRC8 (Q96P09) | TPPP3 (Q9BW30) | VCAM1 (P19320) | ||
| CASP8 (Q14790) | TTBK1 (Q5TCY1) | |||
| CASP9 (P55211) | TTBK2 (Q61Q55) | |||
| Alzheimer Related | Cartilage Related | AMD Related Proteins | ECM Proteins | |
| Total Hits 4 | Total Hits 14 | Total Hits 4 | Total Hits 40 | |
| APP (P05067) | CHAD (Q15339) | BEST1 (O76090) | BSG (P35613) | |
| BPTF (Q12830) | COMP (P49747) | BEST2 (Q89FU1) | CD44 (P16070) | |
| COL25A1 (Q9BXS0) | GDF5 (P43026) | BEST3(Q8N1M1) | ECM1 (Q16610) | |
| CLSTN1(O94985) | MATN1 (P21941) | BEST4(Q8NFU0) | ELFN1 (P0C7U0) | |
| BGN (P21810) | TNC (P24821) | |||
Accession numbers are shown in parentheses next to each protein.
Search motif used to produce this partial list is as follow: [AEGR]-[LAGR]-[IAMLR]-[TVIAL].
For MDR proteins, # of accessible sites (outside of transmembrane domains) and total # of putative cut sites are indicated in the last column.
Identification of tubulins as substrates of HtrA1
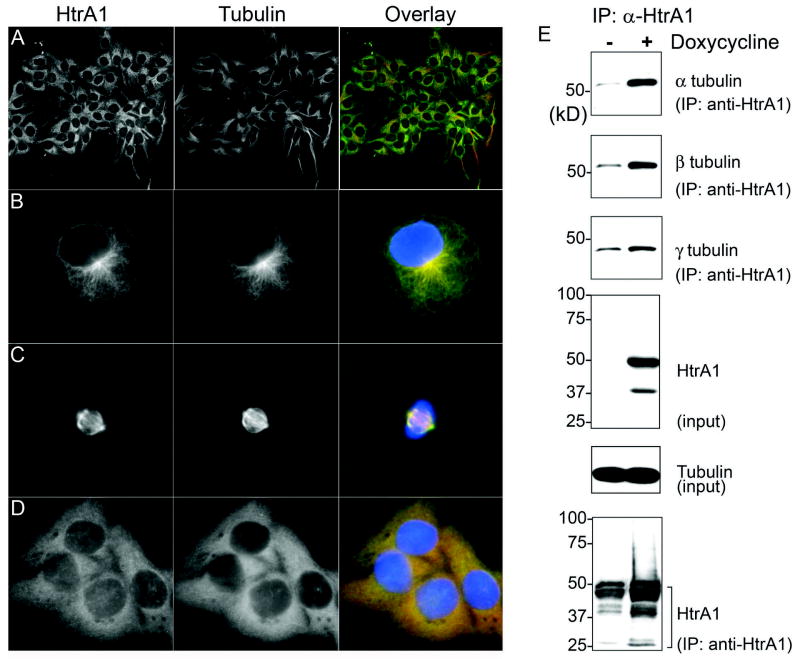
A set of the proteins identified as potential substrates of HtrA1 included tubulins (Table 1). These results were consistent with our initial independent observation that HtrA1 localizes to microtubules (Fig. 3A-D). HtrA1 is localized to microtubules in both interphase and mitotic cells (Fig. 3B-C). In addition, destabilization of microtubules by Nocodazole (5 μM for 2 hours) resulted in diffuse staining of both tubulin and HtrA1 indicating that cytoskeletal localization of HtrA1 is dependent on intact microtubules (Fig. 3D). Finally, in 293T cell line with inducible expression of HtrA1, immunoprecipitation of HtrA1 resulted in co-precipitation of all three types of tubulins (Fig 3E). These results further support our analysis from peptide library screenings that tubulins may serve as potential substrates of HtrA1. Therefore, we selected tubulins for further analyses to determine whether they can be degraded by HtrA1.
Fig. 3.
Localization of endogenous HtrA1 to microtubules. A. Endogenous expression of HtrA1 in SKOV3 cells (observed under 10x objective lens). B-C. Localization of HtrA1 to microtubules at interphase and mitotic cells. Overlay indicates merging of pseudocolors for HtrA1 (green), tubulin (yellow), and nuclear stain DAPI (blue). D. Tubular staining of HtrA1 was dependent on intact microtubules, and destabilization of microtubules by Nocodazole treatment resulted in diffuse staining of HtrA1. E. In 293T cells with inducible HtrA1 expression, immunoprecipitation of HtrA1 resulted in co-precipitation of α-, β-, and γ-tubulins.
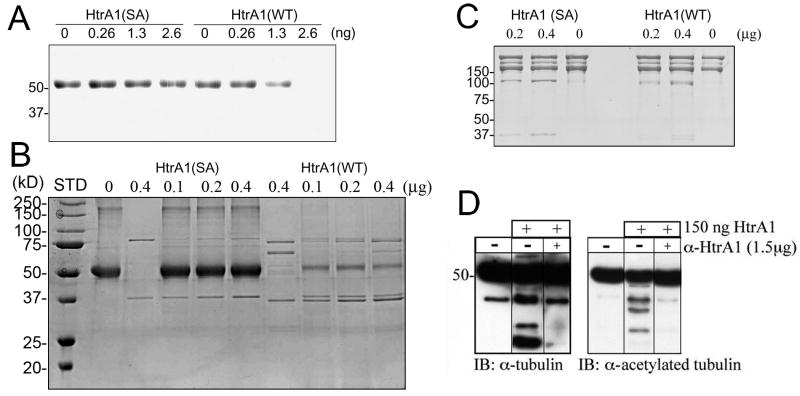
HtrA1 degrades purified tubulins in vitro
To determine whether HtrA1 degrades tubulins, various concentrations of purified HtrA1 were incubated for 3 h with 500 ng of bovine brain-purified tubulins, and reaction products were resolved by SDS-PAGE. Coomassie blue staining of gels indicates dose-dependent degradation of tubulins by wild type HtrA1 (Fig. 4A). In contrast, protease mutant (SA) did not show tubulin degradation, indicating that it is dependent on HtrA1 protease activity. We could not detect partially degraded products of tubulins by Coomassie stain (Fig. 4A). Because we started with very small amount of tubulins in the digestion reaction, it is possible that partially degraded tubulins could be below the detection limit of Coomassie stain. Therefore, we next started with 10 μg of tubulins in the subsequent digestion experiments with higher amounts of HtrA1 as well. However, we did not observe partially degraded products of HtrA1 (Fig. 4B). Only when we performed immunoblotting with anti-tubulin antibodies, we were able to detect partially degraded tubulins in the digestion reactions (Fig. 4D). Under similar conditions, we did not observe degradation of type I collagens (Fig. 4C), demonstrating the selectivity of HtrA1 toward specific substrates.
Fig. 4.
In vitro degradation of tubulins by HtrA1. A. Dose dependent degradation of tubulins by HtrA1. B. No partially digested tubulins were observed by Coomassie stain even when 10 μg of tubulin was used in the digestion reaction. C. No collagen degradation was observed when 2 μg of type I collagen was tested as a potential substrate. D. Partially degraded tubulins were observed only by immunoblotting, and could be blocked by a neutralizing antibody against HtrA1 (α-HtrA1).
HtrA1 degrades tubulins in intact cells
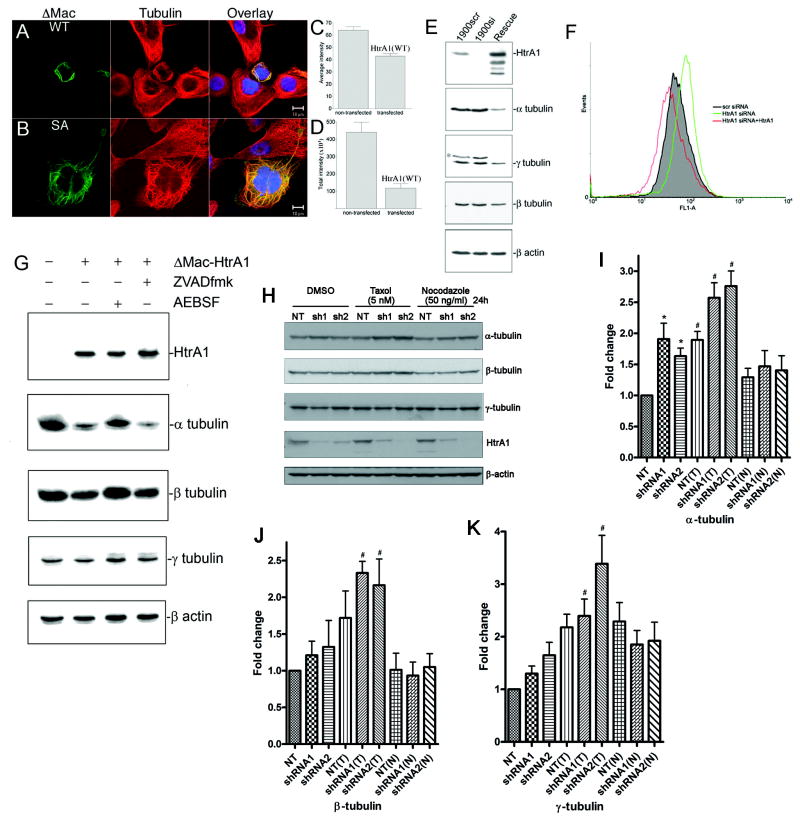
To determine whether HtrA1 could target tubulins in intracellular environment in intact cells, we transfected either wild type or protease mutant HtrA1 into ovarian cancer cell line, OV202, and immunolocalization of HtrA1 and immunofluorescence intensity of α-tubulin were determined. Following expression of GFP-tagged HtrA1, it was observed to be co-localized with microtubules. In addition, transfected cells displayed shrinkage of cytoplasmic volume, rounded cell morphology, and reduction in a-tubulin immunofluorescence (Fig. 5A). These changes in cell morphology and tubulin fluorescence are dependent on the protease activity of HtrA1 because these changes are not observed in cells transfected with the protease mutant HtrA1 (Fig. 5B). Quantitative analyses of tubulin immunofluorescence intensity within transfected cells compared to neighboring non-transfected cells also indicate a decrease in tubulin immunofluorescence within transfected cells (Fig. 5C-D). Similarly, in SKOV3 cells with endogenous HtrA1, down-regulation of HtrA1 by RNAi resulted in slight but noticeable increased immunoreactivity to α- and γ-tubulin. Most significantly, rescue of HtrA1 expression by ectopic expression of HtrA1 resulted in decreased immunoreactivity to α-tubulins and γ-tubulins (Fig. 5E). Flow cytometry analysis also indicates an increase in α-tubulin immunofluorescence following down-regulation of HtrA1, but a decrease in α-tubulin immunofluorescence following rescue of HtrA1 expression (Fig. 5F). Finally, in OV202 cell line with no detectable endogenous HtrA1 expression, forced expression of HtrA1 reduces the endogenous levels of α- and β-tubulins. This reduction in endogenous tubulin levels can be rescued by pre-incubation of cells with serine protease inhibitor, AEBSF, but not with broad caspase inhibitor, ZVADfmk, demonstrating that decreased endogenous levels of tubulins were as a result of HtrA1 protease activities (Fig. 5G).
Fig. 5.
Degradation of intracellular tubulins by HtrA1 in intact cells. A. Exogenous expression of wild type HtrA1 (WT) in OV202 resulted in reduced tubulin immunofluorescence. B. Exogenous expression of protease mutant HtrA1 (SA) resulted in bundling of microtubules. C. Tubulin immunofluorescence intensity analysis indicates average intensity of tubulins within transfected cells decreased compared to non-transfected cells when adjusted for surface area of the cell. D. Overall intensity of tubulins, not adjusted for surface area, decreased markedly in transfected cells compared to non-transfected neighboring cells. E. In SKOV3 cells with endogenous HtrA1, down-regulation of HtrA1 by siRNA minimally increases endogenous tubulins, and rescue of HtrA1 expression in these cells markedly decreases endogenous tubulins. F. Flow cytometry analysis of α-tubulin expression in these cells shows a decrease in tubulin immunofluorescence following rescue of HtrA1 expression, whereas an increase in tubulin immunofluorescence is observed when endogenous HtrA1 is down-regulated by siRNA. G. Decreased in endogenous tubulins following HtrA1 expression is dependent on serine protease activities and could be inhibited by serine protease inhibitor, AEBSF, but not by broad caspase inhibitor ZVADfmk. H. Alpha tubulin levels are higher in SKOV3 cells with stable knockdown of HtrA1 by two different shRNAs (shRNA1 and shRNA2) than in cells with non-targeting shRNA (NT). Levels of α-, β-, and γ-tubulins are higher in SKOV3 cells treated for 24 hours with 5 nM paclitaxel (Taxol) than in cells treated with vehicle (DMSO) or with 50 ng/ml Nocodazole (top 3 panels). HtrA1 is efficiently knocked down in SKOV3 cells by shRNA1 and shRNA2 targeting HtrA1 (4th panel from top). β-actin immunoblot indicates controls for protein loading. I-K. Densitometry analysis of tubulin expression in SKOV3 cells with stable transfection of shRNA following 24 hours treatment with agents that altered microtubule stability or vehicle control. Expression values are normalized with β-actin expression and indicated as fold-change over control (DMSO-treated SKOV3 with stable transfection of non-targeting shRNA). (T) indicates Taxol treatment, and (N) indicates Nocodazole treatment. Bar graphs represent mean±sem; * p<0.05, with Newman-Keuls Multiple Comparison Test; # p<0.05 with Dunnett’s Multiple Comparison Test.
Effect of HtrA1 on tubulin levels under agents that modulate microtubule stability
To determine whether stable knock-down of HtrA1 has an effect on tubulin levels in SKOV3 cells, we stably infected the cells with lentiviruses containing HtrA1-targeting shRNAs (shRNA1 and shRNA2) and non-targeting shRNA (NT) as a control. Batch-stable cell lines were generated and treated with Taxol, Nocodazole, or DMSO vehicle for 24 hours. Immunoblot analyses indicate higher levels of α-tubulins in SKOV3 cells with stable knock-down of HtrA1 (Fig. 5H). Densitometric analyses from three independent experiments indicate a statistically significant increase in α-tubulin levels following stable knock-down HtrA1 by two different shRNAs (Fig. 5I). Although we also observed the increase in α-tubulin levels in HtrA1 knock-down cells following Taxol treatments (Fig. 5H), the increase was not statistically significant (Fig. 5I). No significant increase in β-tubulin or γ-tubulin levels was observed following HtrA1 knock-down (Fig. 5H, 5J, 5K). Consistent with previous studies [Gong and Brandhorst, 1988], stabilization of microtubules by Taxol significantly increases all three forms of tubulins (Fig. 5, H-K). On the other hand, destabilization of microtubules by Nocodazole has no effect on overall levels of tubulins (Fig. 5, H-K). These results further confirm the potential role of endogenous HtrA1 in regulating the levels of α-tubulin.
DISCUSSION
Serine protease HtrA1 has been implicated in pathogenesis of various diseases, including age-related macular degeneration, Alzheimer’s disease, osteoarthritis, preeclampsia, and tumor progression [Ajayi et al., 2008; Baldi et al., 2002; Chien et al., 2006; Chien et al., 2004; Grau et al., 2005; Grau et al., 2006; Hu et al., 1998; Tsuchiya et al., 2005; Yang et al., 2006]. However, little is known about biological (pathologic or physiologic) substrates of HtrA1 or the role of these substrates in pathogenesis associated with HtrA1. In this study, our effort to identify potential substrates of HtrA1 by determination of HtrA1 cleavage motif produced many promising candidates that need to be further validated.
We used mixture-based oriented peptide library approach to identified potential substrates of HtrA1. This approach has been successfully used by others to identify potential substrates of matrix metalloproteinases and calpain [Cuerrier et al., 2005; Turk et al., 2001]. For example, Turk et al had successfully used this approach to identify neurocan as a substrate of MMP-2 [Turk et al., 2001]. In addition, Turk et al, in the same study, identified cleavage site motifs of six MMPs. They observed poor selectivity of P’ residues, with the exception of P1’ of cleavage site motifs for some MMPs. Unlike MMPs, our results suggest that HtrA1 serine protease has some selectivity to both primed and unprimed cleavage motif. The primary sequence specificity of unprimed cleavage sites indicates that the protease favors cleavage following aliphatic residues, similar to bacterial HtrA1 and consistent with sequence specificity of mammalian HtrA2 [Martins et al., 2003]. However, unlike HtrA2 that shows preference for aliphatic residues at P1 and P4 [Martins et al., 2003], HtrA1 shows preference for aliphatic residues at all four unprimed positions (with the exception of polar threonine residue at P1) (Fig. 2). For the primed positions, the protease shows preference to hydrophilic polar residues at P1’ and P3’ (Fig. 1). In addition, compared to the unprimed position, more defined primary sequence specificity was observed for residues at primed positions (Fig. 1). Similar approach has been used by Cuerrier et al to show the primed side selectivity of μ-calpain [Cuerrier et al., 2005].
In this study, we identified tubulins as potential substrates of HtrA1 and further characterized these proteins as in vitro and intracellular substrates of HtrA1. The fact that HtrA1 co-localizes to intracellular microtubules is also consistent with the observation that tubulins may serve as potential substrates of HtrA1. Moreover, we observed co-immunoprecipitation of α-, β-, and γ- tubulins when endogenous HtrA1 is immunoprecipitated (data not shown). In addition, sedimentation of microtubules by ultracentrifugation resulted in co-sedimentation of HtrA1 in microtubules pellets (data not shown). Finally, in vitro and intracellular tubulin degradation studies further support the role of tubulins as in vitro and intracellular substrates of HtrA1. Although several proteases, such as calpains, caspases, granzyme B and granzyme M, have been implicated in degradation of tubulins in vitro [Adrain et al., 2006; Bovenschen et al., 2008; Goll et al., 2003], we are the first to report serine protease HtrA1 targeting tubulins for in vitro and intracellular degradation. It is interesting to note that levels of intracellular α- and β-tubulins decreased following overexpression of HtrA1, and that this decrease in α- and β-tubulin levels was prevented by pre-incubating the cells with serine protease inhibitor, AEBSF, but not by broad caspase inhibitor, ZVADfmk. These results support the role of HtrA1 and other serine proteases in targeting tubulins for degradation.
It is important to note that our observation that tubulins may serve as potential substrates of HtrA1 should be viewed in the pathophysiologic context. Although our knockdown experiments, using transient and stable knock-down of HtrA1, provided the evidence for a role of endogenous HtrA1 in regulating α-tubulin levels, the physiologic significance of such regulation is not currently known. Nonetheless, we showed that over-expression of HtrA1 contributed to tubulin degradation and microtubule loss. Previous studies have shown that HtrA1 is upregulated and activated during chemotherapy-induced cytotoxicity [Chien et al., 2006]. Moreover, HtrA1 is upregulated in osteoarthritis and in preeclampsia [Ajayi et al., 2008; Grau et al., 2006; Tsuchiya et al., 2005]. Therefore, our observation that HtrA1 may target tubulins for degradation should be viewed in the pathophysiologic contexts whereby aberrant HtrA1 expression may contribute to alterations in cytoskeletal dynamics that accompanied the initiation of cell death programs or pathogenic processes such as Alzheimer’s disease, osteoarthritis, or preeclampsia. Under these pathologic conditions, it will be interesting to determine whether pathologic substrates of HtrA1 contribute to pathogenesis of these diseases.
It is also important to note that HtrA1 may have different roles in physiologic and pathologic conditions. For example, full-length HtrA1 contains N-terminal Kazal-type trypsin inhibitor that may serve as intra- or inter-molecular inhibitor of HtrA protease activity. In fact, our previous studies indicate that full-length HtrA1 is less potent inducer cell death compare to N-terminal deleted HtrA1, suggesting that N-terminal Kazal-inhibitor serves to limit protease activity of HtrA1 [Chien et al., 2006]. Therefore, it is possible that intact HtrA1 may serve as microtubule-associated protein and regulate cellular functions related to microtubules, rather than as a protease that targets tubulins for degradation under physiologic conditions. However, under pathologic conditions, whereby its homeostasis is altered by aberrant expression of HtrA1 or its regulatory proteins, HtrA1 may target tubulins for degradation and contribute to the pathogenic processes. For example, under conditions of stress by chemotherapeutic agents and upon N-terminal cleavage of HtrA1 during programmed cell death [Chien et al., 2006], active HtrA1 may target tubulins and microtubules and contribute to cell death. This function will be consistent with its pro-apoptotic properties reported in our previous studies [Chien et al., 2006; Chien et al., 2004]. In addition, HtrA1, under physiologic condition, may target mis-folded tubulins for degradation, and thereby may promote incorporation of properly folded tubulins to enhance polymerization of microtubules. Since microtubules are important in many cellular functions, including cell migration, cell division, cell shape integration, and organelle and protein transport, it will be important to investigate the potential role of HtrA1 in these cellular functions.
Our evidence also indicates that enhanced expression of HtrA1 in ovarian cancer cell line OV202 resulted in microtubule loss as evidence by decreased in intact microtubules and immunostaining intensity. This microtubule severing activity is dependent on protease activity of HtrA1 because no microtubule loss was observed with overexpression of protease mutant HtrA1. Interestingly, overexpression of protease mutant HtrA1 resulted in bundling of microtubules, suggesting that HtrA1 may function as a microtubule-associated protein. Similarly, calpain 6, a member of cysteine protease family, with no apparent protease activity also associates with microtubules and causes microtubule bundles when overexpressed in cells [Tonami et al., 2007]. These results suggest a role of proteases in regulating microtubule dynamics, cytoskeletal reorganization, and tubulin turnover.
The fact that HtrA1 antibodies targeted to the PDZ domain of HtrA1 partially blocked degradation of tubulins by HtrA1 suggests the potential role of the PDZ domain as a substrate recognition module. This result is consistent with previous reports indicating that PDZ domains of HtrA proteins may serve as substrate recognition and regulator of protease activities [Li et al., 2002; Murwantoko et al., 2004; Runyon et al., 2007; Schlieker et al., 2004; Sohn et al., 2007]. In particular, previous studies by Runyon et al reported the binding specificity of PDZ domain of HtrA1. Because PDZ domain of HtrA1 protein is considered as a substrate recognition domain, the binding specificity infers substrate specificity. However, it is important to note that while Runyon et al focuses on binding specificity of PDZ domain, our study focuses on cleavage specificity of protease domain. It is likely that both of these specificities contribute to substrate specificity. It is possible that HtrA1 may bind to a substrate through PDZ domain in accordance with the binding specificity given by PDZ domain, but may cleave the substrate at specific site based on specificity given by protease domain. Therefore, differences in sequence specificity between these studies reflect differences in specificity of PDZ domain and protease domain. Previous reports by Martins et al [Martins et al., 2003] and Oka et al [Oka et al., 2004] did indicate that protease domain possess inherent specificity of candidate substrates. Runyon et al and others have reported that specificity may also be conferred by PDZ domain. In addition, HtrA1, HtrA3, and HtrA4 contain N-terminal Mac25 domain, and therefore, it is possible that Mac25 domain may also play a role in regulating substrate specificity [Runyon et al., 2007]. Consequently, it is critical to dissect the specificities of the individual domains and determine how these domains function together to contribute to substrate specificity [Runyon et al., 2007]. Therefore, both Runyon et al and our studies contribute to increased understanding of substrate specificity inherent in HtrA1 protein. In this context, the identification of promiscuous hydrophobic residues as binding motif of PDZ domain of HtrA1 by Runyon et al is quite informative and is consistent with its putative role in the recognition of unfolded proteins. It is possible that for denatured substrates, PDZ domain a play a critical role in the recognition of these unfolded protein with specificity provided by PDZ domain binding to hydrophobic residues. However, it is unlikely that the recognition and degradation of unfolded proteins by HtrA1 through PDZ is the only role for this protease. Evidence suggests that HtrA1 associates with TGF-β and regulates its signaling through the protease domain. HtrA1 also binds to IGFBP5 through its Mac25 domain and regulates its degradation. Recently, several junctional proteins and ion channels have been identified to bind to PDZ domain of HtrA1 and HtrA3 in very specific manner [Stiffler et al., 2007]. These results therefore suggest a more complex role of these proteases, in addition to their role in protein quality control.
The search of potential substrates using HtrA1 cleavage motifs also produces various proteins that were previously suggested to be candidate substrates for HtrA1. For example, collagens, fibromodulins, fibronectin, aggrecan, decorin were previously identified as substrates of HtrA1 [Grau et al., 2006; Tsuchiya et al., 2005]. Our cleavage motif scan also indicates these proteins as potential substrates of HtrA1 (Table 1). Amyloid precursor protein has been previously shown to be a substrate of HtrA1 [Grau et al., 2005]. Our analysis also identified APP as a substrate of HtrA1, and therefore is consistent with previous report [Grau et al., 2005]. In addition, our analysis identified several putative substrates that may play a role in various disease pathologies, such as age-regulated macular degeneration, arthritis, and Alzheimer’s disease. However, abundant caution needs to be applied in selecting these candidate substrates for further validation since the list was generated with the most inclusive search criteria involving at least four degenerate amino acids for each unprimed positions corresponding to P1 to P4. It is recommended that much more restrictive search criteria involving just two most selective amino acids for each position should be applied in selecting candidate substrates for further validation.
Other potential groups of HtrA1 substrates include inhibitor of apoptosis proteins (IAPs). In particular, XIAP has been identified as a potential substrate of HtrA1. It is interesting to note that IAPs have been identified as substrates of HtrA2, a second member of mammalian HtrA family. Therefore, it is tempting to speculate that HtrA serine proteases may share overlapping substrates. It is important to note that both HtrA1 and HtrA2 are described as pro-apoptotic proteins. In addition, expression of HtrA1 modulates better response to chemotherapy in vitro and associates with better response to chemotherapy in vivo [Chien et al., 2006]. Interestingly, enhanced expression of HtrA1 resulted in increased activities of caspase 3/7 [Chien et al., 2006]. Since IAPs regulate caspase activities, it is conceivable that HtrA1 may regulate caspase activities by regulating IAPs.
Previous studies have shown that enhanced expression of either HtrA1 or HtrA2 resulted in cells with rounded cell morphology [Chien et al., 2004; Suzuki et al., 2001]. In light of the current finding that HtrA1 may target tubulins for degradation, it is tempting to speculate that HtrA1 may disrupt microtubules by targeting tubulins, resulting in cell rounding and cell death. It will also be important to investigate whether HtrA1 plays any role in cytoskeletal remodeling during chemotherapy-induced cell death. Our analysis also identified several cytoskeletal and adhesion proteins as potential substrate of HtrA1. Therefore, it is likely that targeted degradation of these potential substrates may play a role in cytoskeletal remodeling during cell death programs.
Finally, our analysis also points to various transporter proteins as potential substrates of HtrA1. These candidate substrates include MDR1, MDR3, and ABCB proteins. Since these proteins contain multiple transmembrane domains, which are unlikely to be accessible by the protease, we indicated potential accessible cut sites (outside of transmembrane domains) as well as all cut sites identified by primary sequence. These proteins play an important role in resistance to chemotherapy in vitro. Therefore, it will be of significant interest to investigate the extent to which expression and function of these transporter proteins are altered as a result of HtrA1 expression. These studies will undoubtedly provide additional functional understanding of the role of HtrA1 in various pathobiologic processes.
Supplementary Material
In vitro collagen degradation assays. Type III and IV collagens were used as potential substrates of HtrA1. No degradation of collagens was observed in vitro. Coomassie stained protein band corresponding to purified HtrA1 is indicated by text next to the gel image.
Acknowledgments
This work was supported by grants from National Cancer Institute, 1R01CA123249 (to VS and JC), the Mayo Clinic Bernard and Edith Waterman Center for Cancer Genetics and the Mayo Foundation (to V.S.) and from Ovarian Cancer Research Fund (to JC).
References
- Adrain C, Duriez PJ, Brumatti G, Delivani P, Martin SJ. The cytotoxic lymphocyte protease, granzyme B, targets the cytoskeleton and perturbs microtubule polymerization dynamics. J Biol Chem. 2006;281:8118–25. doi: 10.1074/jbc.M509361200. [DOI] [PubMed] [Google Scholar]
- Ajayi F, Kongoasa N, Gaffey T, Asmann YW, Watson W, Baldi A, Lala P, Shridhar V, Brost B, Chien J. Elevated expression of serine protease HtrA1 in preeclampsia and its role in trophoblast cell migration and invasion. American Journal of Obstetrics and Gynecology. 2008 doi: 10.1016/j.ajog.2008.04.046. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Baldi A, De Luca A, Morini M, Battista T, Felsani A, Baldi F, Catricala C, Amantea A, Noonan DM, Albini A, Natali PG, Lombardi D, Paggi MG. The HtrA1 serine protease is down-regulated during human melanoma progression and represses growth of metastatic melanoma cells. Oncogene. 2002;21:6684–8. doi: 10.1038/sj.onc.1205911. [DOI] [PubMed] [Google Scholar]
- Bovenschen N, de Koning PJ, Quadir R, Broekhuizen R, Damen JM, Froelich CJ, Slijper M, Kummer JA. NK cell protease granzyme M targets alpha-tubulin and disorganizes the microtubule network. J Immunol. 2008;180:8184–91. doi: 10.4049/jimmunol.180.12.8184. [DOI] [PubMed] [Google Scholar]
- Chien J, Aletti G, Baldi A, Catalano V, Muretto P, Keeney GL, Kalli KR, Staub J, Ehrmann M, Cliby WA, Lee YK, Bible KC, Hartmann LC, Kaufmann SH, Shridhar V. Serine protease HtrA1 modulates chemotherapy-induced cytotoxicity. J Clin Invest. 2006;116:1994–2004. doi: 10.1172/JCI27698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien J, Staub J, Hu SI, Erickson-Johnson MR, Couch FJ, Smith DI, Crowl RM, Kaufmann SH, Shridhar V. A candidate tumor suppressor HtrA1 is downregulated in ovarian cancer. Oncogene. 2004;23:1636–44. doi: 10.1038/sj.onc.1207271. [DOI] [PubMed] [Google Scholar]
- Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell. 2002;10:443–55. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- Clawson GA, Bui V, Xin P, Wang N, Pan W. Intracellular localization of the tumor suppressor HtrA1/Prss11 and its association with HPV16 E6 and E7 proteins. J Cell Biochem. 2008;105:81–8. doi: 10.1002/jcb.21804. [DOI] [PubMed] [Google Scholar]
- Cuerrier D, Moldoveanu T, Davies PL. Determination of peptide substrate specificity for mu-calpain by a peptide library-based approach: the importance of primed side interactions. J Biol Chem. 2005;280:40632–41. doi: 10.1074/jbc.M506870200. [DOI] [PubMed] [Google Scholar]
- De Luca A, De Falco M, Severino A, Campioni M, Santini D, Baldi F, Paggi MG, Baldi A. Distribution of the serine protease HtrA1 in normal human tissues. J Histochem Cytochem. 2003;51:1279–84. doi: 10.1177/002215540305101004. [DOI] [PubMed] [Google Scholar]
- Faccio L, Fusco C, Chen A, Martinotti S, Bonventre JV, Zervos AS. Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J Biol Chem. 2000;275:2581–8. doi: 10.1074/jbc.275.4.2581. [DOI] [PubMed] [Google Scholar]
- Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- Gong ZY, Brandhorst B. Autogenous regulation of tubulin synthesis via RNA stability during sea urchin embryogenesis. Development. 1988;102:31–43. doi: 10.1242/dev.102.1.31. [DOI] [PubMed] [Google Scholar]
- Grau S, Baldi A, Bussani R, Tian X, Stefanescu R, Przybylski M, Richards P, Jones SA, Shridhar V, Clausen T, Ehrmann M. Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proc Natl Acad Sci U S A. 2005;102:6021–6. doi: 10.1073/pnas.0501823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau S, Richards PJ, Kerr B, Hughes C, Caterson B, Williams AS, Junker U, Jones SA, Clausen T, Ehrmann M. The role of human HtrA1 in arthritic disease. The Journal of biological chemistry. 2006;281:6124–9. doi: 10.1074/jbc.M500361200. [DOI] [PubMed] [Google Scholar]
- Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D, Southan C, Barton A, Fantom KG, West A, Savopoulos J, Hassan NJ, Clinkenbeard H, Hanning C, Amegadzie B, Davis JB, Dingwall C, Livi GP, Creasy CL. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur J Biochem. 2000;267:5699–710. doi: 10.1046/j.1432-1327.2000.01589.x. [DOI] [PubMed] [Google Scholar]
- Hu SI, Carozza M, Klein M, Nantermet P, Luk D, Crowl RM. Human HtrA, an evolutionarily conserved serine protease identified as a differentially expressed gene product in osteoarthritic cartilage. J Biol Chem. 1998;273:34406–12. doi: 10.1074/jbc.273.51.34406. [DOI] [PubMed] [Google Scholar]
- Li W, Srinivasula SM, Chai J, Li P, Wu JW, Zhang Z, Alnemri ES, Shi Y. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat Struct Biol. 2002;9:436–41. doi: 10.1038/nsb795. [DOI] [PubMed] [Google Scholar]
- Lingle WL, Salisbury JL. Methods for the analysis of centrosome reproduction in cancer cells. Methods Cell Biol. 2001;67:325–36. doi: 10.1016/s0091-679x(01)67022-5. [DOI] [PubMed] [Google Scholar]
- Lipinska B, Sharma S, Georgopoulos C. Sequence analysis and regulation of the htrA gene of Escherichia coli: a sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1988;16:10053–67. doi: 10.1093/nar/16.21.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins LM, Turk BE, Cowling V, Borg A, Jarrell ET, Cantley LC, Downward J. Binding specificity and regulation of the serine protease and PDZ domains of HtrA2/Omi. J Biol Chem. 2003;278:49417–27. doi: 10.1074/jbc.M308659200. [DOI] [PubMed] [Google Scholar]
- Murwantoko, Yano M, Ueta Y, Murasaki A, Kanda H, Oka C, Kawaichi M. Binding of proteins to the PDZ domain regulates proteolytic activity of HtrA1 serine protease. Biochem J. 2004;381:895–904. doi: 10.1042/BJ20040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie GY, Hampton A, Li Y, Findlay JK, Salamonsen LA. Identification and cloning of two isoforms of human high-temperature requirement factor A3 (HtrA3), characterization of its genomic structure and comparison of its tissue distribution with HtrA1 and HtrA2. Biochem J. 2003;371:39–48. doi: 10.1042/BJ20021569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka C, Tsujimoto R, Kajikawa M, Koshiba-Takeuchi K, Ina J, Yano M, Tsuchiya A, Ueta Y, Soma A, Kanda H, Matsumoto M, Kawaichi M. HtrA1 serine protease inhibits signaling mediated by Tgfbeta family proteins. Development. 2004;131:1041–53. doi: 10.1242/dev.00999. [DOI] [PubMed] [Google Scholar]
- Pallen MJ, Wren BW. The HtrA family of serine proteases. Mol Microbiol. 1997;26:209–21. doi: 10.1046/j.1365-2958.1997.5601928.x. [DOI] [PubMed] [Google Scholar]
- Runyon ST, Zhang Y, Appleton BA, Sazinsky SL, Wu P, Pan B, Wiesmann C, Skelton NJ, Sidhu SS. Structural and functional analysis of the PDZ domains of human HtrA1 and HtrA3. Protein Sci. 2007;16:2454–71. doi: 10.1110/ps.073049407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Mogk A, Bukau B. A PDZ switch for a cellular stress response. Cell. 2004;117:417–9. doi: 10.1016/s0092-8674(04)00453-2. [DOI] [PubMed] [Google Scholar]
- Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131:572–83. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–47. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–9. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch KL, Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci U S A. 1988;85:1576–80. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–21. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- Tonami K, Kurihara Y, Aburatani H, Uchijima Y, Asano T, Kurihara H. Calpain 6 is involved in microtubule stabilization and cytoskeletal organization. Mol Cell Biol. 2007;27:2548–61. doi: 10.1128/MCB.00992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya A, Yano M, Tocharus J, Kojima H, Fukumoto M, Kawaichi M, Oka C. Expression of mouse HtrA1 serine protease in normal bone and cartilage and its upregulation in joint cartilage damaged by experimental arthritis. Bone. 2005;37:323–36. doi: 10.1016/j.bone.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Turk BE, Cantley LC. Using peptide libraries to identify optimal cleavage motifs for proteolytic enzymes. Methods. 2004;32:398–405. doi: 10.1016/j.ymeth.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat Biotechnol. 2001;19:661–7. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, Chien J, Dewan A, Harmon J, Bernstein PS, Shridhar V, Zabriskie NA, Hoh J, Howes K, Zhang K. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–3. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- Zumbrunn J, Trueb B. Primary structure of a putative serine protease specific for IGF-binding proteins. FEBS Lett. 1996;398:187–92. doi: 10.1016/s0014-5793(96)01229-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vitro collagen degradation assays. Type III and IV collagens were used as potential substrates of HtrA1. No degradation of collagens was observed in vitro. Coomassie stained protein band corresponding to purified HtrA1 is indicated by text next to the gel image.