Abstract
The title compound, C21H21N3O2, was obtained following a five-step synthetic procedure yielding weakly diffracting rod and needle-shaped crystals which crystallized concomitantly. Structural analysis of a rod-shaped crystal showed that the central seven-membered heterocyclic ring adopts a conformation that is perhaps best described as a distorted boat, with the H-bearing (CH2 and NH) atoms lying well out of the least-squares mean plane fitted through the other five atoms in the ring (r.m.s. deviation 0.075 Å). In the crystal, the compound packs as a twisted chain, which propagates along the b axis by means of an R 1 2(6) motif formed by one of the carbonyl O atoms acting as a bifurcated acceptor in an N—H⋯O and C—H⋯O interaction. No diffraction was observed from the needle-shaped crystals.
Related literature
For background to the synthetic procedure, see: Hulme & Gore (2003 ▶); Hulme et al. (2000 ▶). For graph-set analysis of hydrogen-bond networks, see: Bernstein et al. (1995 ▶).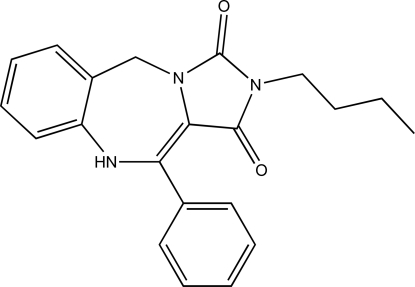
Experimental
Crystal data
C21H21N3O2
M r = 347.41
Monoclinic,
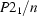
a = 12.192 (4) Å
b = 7.638 (2) Å
c = 18.514 (6) Å
β = 95.494 (5)°
V = 1716.1 (9) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 100 K
0.29 × 0.14 × 0.08 mm
Data collection
Bruker Kappa APEXII DUO CCD diffractometer
Absorption correction: numerical (SADABS; Sheldrick, 1996 ▶) T min = 0.975, T max = 0.993
14047 measured reflections
2724 independent reflections
1908 reflections with I > 2σ(I)
R int = 0.067
θmax = 24.1°
Refinement
R[F 2 > 2σ(F 2)] = 0.065
wR(F 2) = 0.181
S = 1.03
2724 reflections
239 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.71 e Å−3
Δρmin = −0.29 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: SHELXTL and local programs.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810005477/bh2272sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810005477/bh2272Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H3N⋯O1i | 0.86 (4) | 2.10 (4) | 2.944 (3) | 165 (3) |
| C8—H8⋯O1i | 0.95 | 2.57 | 3.326 (4) | 136 |
Symmetry code: (i) 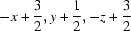 .
.
Acknowledgments
The diffractometer was purchased with funding from NSF grant No. CHE-0741837.
supplementary crystallographic information
Comment
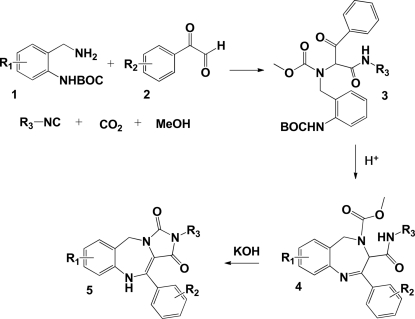
We recently investigated a three step solution phase protocol for the synthesis of arrays of tricyclic fused hydantoin-benzodiazepines as part of broader research on multi-component reactions (Figure 1). Interestingly, the major product of this unique synthetic route was the tautomer 5 derived from the originally desired product 4, the structure being confirmed by X-ray crystallography. The methodology employs ortho-N-Boc benzylamines 1 and phenylglyoxaldehydes 2 in the rarely used five component Ugi reaction with CO2 to assemble desired diversity in product 3 (Hulme et al., 2000). Acid treatment unmasks an internal amino nucleophile and promotes rapid formation of the diazepine ring of generic structure 4. Subsequent base treatment employs the amidic NH of the Ugi scaffold as a second internal nucleophile promoting hydantoin formation and an unexpected 1,3-H shift to give 5. As such the methodology represents an example of a post-condensation Ugi modification (Hulme & Gore, 2003) that employs two internal nucleophiles in distinct operations, generating a novel scaffold of high complexity in three succinct functional operations.
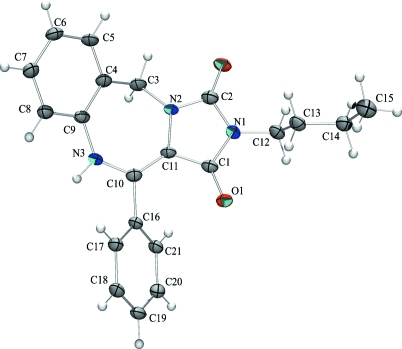
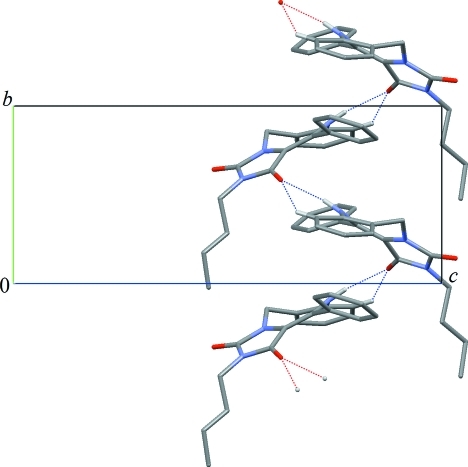
Two types of crystals were formed: very fine yellow needles together with a few slightly larger rod-shaped pale yellow crystals. The needles did not give any measurable diffraction and the rod crystals showed weak diffraction with 60 second exposure times; a resolution cutoff of 0.87Å was applied to the dataset. The identity of the needle crystals was not established. The molecular structure of 5 is shown in Figure 2. Molecular dimensions are unexceptional. The amine hydrogen atom was located in a difference Fourier map and its presence is confirmed by participation in hydrogen bonding discussed below. The central 7-membered heterocyclic ring adopts a conformation that is perhaps best described as a distorted boat with the H-bearing (C3 and N3) atoms lying well out of a least squares mean plane fitted through the other five atoms in the ring [r.m.s. deviation 0.075 Å; C3 deviates by 0.679 (5) Å and N3 deviates by 0.301 (4) Å]. The compound packs as a twisted chain which propagates along the b axis by means of an R12(6) motif (Bernstein et al., 1995) formed by one of the carbonyl oxygen atoms acting as bifurcated acceptor in an N–H···O and C–H···O interaction (Figure 3).
Experimental
Ugi reaction (For R1, R2=H, R3= n-butyl)
CO2 gas was bubbled through a stirring solution of MeOH for 25 minutes to generate methyl carbonic acid. In a separate 25 ml flask, phenyl glyoxal 2 (226 mg, 1.687 mmol) was added to BOC-2-aminobenyzlamine 1 (250 mg, 1.125 mmol). Methyl carbonic acid (10 ml) and N-butyl isonitrile (0.237 ml, 2.252 mmol) were then added to the latter flask. The reaction was stirred at room temperature under an atmosphere of CO2 for 16 h. The solvent was evaporated in vacuo and the crude product purified with a Biotage Isolera4TM system (hexane/EtOAc 10-30%) to afford the Ugi product 3 (218 mg, 0.438 mmol, 39%) as a yellow oil.
De-BOC and Cyclization
3 (0.180 g, 0.362 mmol) was treated with a 5 ml 10% TFA solution in 1,2-dichloroethane which was irradiated in a Biotage InitiatorTM at 80°C for 20 min. The resulting orange solution was washed with 1M NaHCO3 (4 × 2.5 ml) and the organic layer dried (Na2SO4), filtered and evaporated in vacuo. MeOH (1.5 ml), THF (0.75 ml), H2O (0.5 ml) were added to the crude product 4 (0.102 g, 0.269 mmol) followed by a 1 g/1 ml solution of KOH in H2O (0.03 ml). The solution was irradiated at 100°C for 20 min and resultant orange solution partitioned between EtOAc (5 ml) and 1M NaHCO3 (5 ml). The organic layer was dried (Na2SO4), filtered and evaporated in vacuo. Final crude product was purified with a Biotage Isolera4TM (hexane/EtOAc 30%) to afford the final product 5 (0.074 g, 0.214 mmol, 80%) as a yellow solid. FT-ICR calculated for C21H22N3O2 [M+H]+: 348.1707, found: 348.1707.
Refinement
A resolution cutoff of 0.87 Å was applied to the dataset due to unobserved diffraction beyond this point. Nevertheless the N—H hydrogen atom was located in a difference Fourier map and the N—H distance freely refined to 0.86 (4) Å with Uiso(H) = 1.2 Ueq(N). C—H atoms were refined with Uiso(H) = 1.5 Ueq(C) (methyl) or Uiso(H) = 1.2 Ueq(C) (all others) with constrained C—H distances in the range 0.95–0.99 Å. The largest residual peak, 0.7 e.Å-3, is approximately 1.53 Å from C21.
Figures
Fig. 1.
Synthetic route to 5.
Fig. 2.
The molecular structure of 5 with displacement ellipsoids at the 50% probability level.
Fig. 3.
An a-axis projection of 5 showing the twisted hydrogen-bonded chain (blue dotted lines; red dotted lines indicate continuation).
Crystal data
| C21H21N3O2 | F(000) = 736 |
| Mr = 347.41 | Dx = 1.345 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 2525 reflections |
| a = 12.192 (4) Å | θ = 2.2–24.8° |
| b = 7.638 (2) Å | µ = 0.09 mm−1 |
| c = 18.514 (6) Å | T = 100 K |
| β = 95.494 (5)° | Rod, yellow |
| V = 1716.1 (9) Å3 | 0.29 × 0.14 × 0.08 mm |
| Z = 4 |
Data collection
| Bruker Kappa APEXII DUO CCD diffractometer | 2724 independent reflections |
| Radiation source: fine-focus sealed tube with Miracol optics | 1908 reflections with I > 2σ(I) |
| graphite | Rint = 0.067 |
| φ and ω scans | θmax = 24.1°, θmin = 1.9° |
| Absorption correction: numerical (SADABS; Sheldrick, 1996) | h = −14→14 |
| Tmin = 0.975, Tmax = 0.993 | k = −8→5 |
| 14047 measured reflections | l = −21→21 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.065 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.181 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.1146P)2 + 0.6555P] where P = (Fo2 + 2Fc2)/3 |
| 2724 reflections | (Δ/σ)max = 0.001 |
| 239 parameters | Δρmax = 0.71 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
| 0 constraints |
Special details
| Experimental. 1H NMR (300 MHz, CDCl3) δ ppm 0.89 (t, J = 7.2 Hz, 3H), 1.28 (m, 2H), 1.56 (m, 2H), 3.49 (t, J = 7.3 Hz, 2H), 4.98 (s, 2H), 5.95 (s, 1H), 6.89 (d, J = 7.8 Hz, 1H), 7.06 (t, J = 7.2 Hz, 1H), 7.28 (t, J = 7.7 Hz, 1H), 7.34 (d, J = 7.5 Hz, 1H), 7.52 (m, 5H).13C NMR (75 MHz, CDCl3) δ ppm 14.1, 20.5, 30.7, 39.0, 45.7, 109.4, 120.7, 123.9, 126.4, 129.1, 129.6, 129.7, 130.5, 130.6, 134.6, 135.4, 142.5, 153.7, 161.9. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.88328 (17) | 0.0841 (3) | 0.87585 (11) | 0.0267 (6) | |
| O2 | 0.61570 (19) | 0.1425 (3) | 1.03172 (11) | 0.0346 (6) | |
| N1 | 0.7648 (2) | 0.0891 (4) | 0.96528 (13) | 0.0249 (6) | |
| N2 | 0.62702 (19) | 0.2468 (3) | 0.91468 (13) | 0.0221 (6) | |
| N3 | 0.6120 (2) | 0.4044 (4) | 0.76450 (14) | 0.0253 (7) | |
| H3N | 0.625 (3) | 0.466 (5) | 0.7271 (19) | 0.030* | |
| C1 | 0.7968 (3) | 0.1375 (4) | 0.89869 (16) | 0.0243 (8) | |
| C2 | 0.6618 (3) | 0.1580 (5) | 0.97655 (17) | 0.0282 (8) | |
| C3 | 0.5292 (3) | 0.3561 (5) | 0.90814 (17) | 0.0290 (8) | |
| H3A | 0.5519 | 0.4803 | 0.9130 | 0.035* | |
| H3B | 0.4843 | 0.3285 | 0.9485 | 0.035* | |
| C4 | 0.4598 (2) | 0.3329 (5) | 0.83796 (17) | 0.0272 (8) | |
| C5 | 0.3498 (3) | 0.2811 (5) | 0.83711 (18) | 0.0308 (8) | |
| H5 | 0.3222 | 0.2501 | 0.8816 | 0.037* | |
| C6 | 0.2800 (3) | 0.2733 (5) | 0.77408 (18) | 0.0316 (8) | |
| H6 | 0.2060 | 0.2351 | 0.7752 | 0.038* | |
| C7 | 0.3188 (3) | 0.3216 (5) | 0.70925 (18) | 0.0295 (8) | |
| H7 | 0.2707 | 0.3223 | 0.6658 | 0.035* | |
| C8 | 0.4285 (3) | 0.3690 (4) | 0.70802 (17) | 0.0274 (8) | |
| H8 | 0.4551 | 0.4017 | 0.6634 | 0.033* | |
| C9 | 0.4995 (2) | 0.3694 (4) | 0.77089 (17) | 0.0241 (8) | |
| C10 | 0.7038 (2) | 0.3312 (4) | 0.80057 (16) | 0.0239 (8) | |
| C11 | 0.7095 (2) | 0.2491 (4) | 0.86567 (16) | 0.0215 (7) | |
| C12 | 0.8299 (3) | −0.0207 (5) | 1.01778 (16) | 0.0291 (8) | |
| H12A | 0.8060 | 0.0008 | 1.0666 | 0.035* | |
| H12B | 0.9083 | 0.0136 | 1.0189 | 0.035* | |
| C13 | 0.8194 (3) | −0.2160 (5) | 1.00075 (16) | 0.0290 (8) | |
| H13A | 0.7411 | −0.2509 | 0.9997 | 0.035* | |
| H13B | 0.8435 | −0.2381 | 0.9520 | 0.035* | |
| C14 | 0.8874 (3) | −0.3268 (5) | 1.05583 (17) | 0.0297 (8) | |
| H14A | 0.8702 | −0.2937 | 1.1052 | 0.036* | |
| H14B | 0.9666 | −0.3032 | 1.0523 | 0.036* | |
| C15 | 0.8653 (3) | −0.5210 (5) | 1.04422 (19) | 0.0377 (9) | |
| H15A | 0.7890 | −0.5471 | 1.0531 | 0.057* | |
| H15B | 0.9157 | −0.5887 | 1.0779 | 0.057* | |
| H15C | 0.8768 | −0.5525 | 0.9942 | 0.057* | |
| C16 | 0.8047 (2) | 0.3506 (4) | 0.76174 (16) | 0.0231 (7) | |
| C17 | 0.8030 (2) | 0.2989 (5) | 0.68966 (16) | 0.0256 (8) | |
| H17 | 0.7377 | 0.2506 | 0.6654 | 0.031* | |
| C18 | 0.8963 (3) | 0.3177 (5) | 0.65308 (16) | 0.0279 (8) | |
| H18 | 0.8943 | 0.2830 | 0.6037 | 0.033* | |
| C19 | 0.9920 (3) | 0.3863 (5) | 0.68772 (17) | 0.0292 (8) | |
| H19 | 1.0559 | 0.3979 | 0.6625 | 0.035* | |
| C20 | 0.9944 (3) | 0.4382 (4) | 0.75934 (17) | 0.0272 (8) | |
| H20 | 1.0601 | 0.4852 | 0.7835 | 0.033* | |
| C21 | 0.9005 (2) | 0.4217 (4) | 0.79611 (17) | 0.0256 (8) | |
| H21 | 0.9022 | 0.4594 | 0.8451 | 0.031* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0233 (12) | 0.0323 (14) | 0.0259 (12) | 0.0001 (10) | 0.0093 (9) | −0.0012 (10) |
| O2 | 0.0315 (13) | 0.0513 (17) | 0.0230 (12) | −0.0006 (11) | 0.0132 (10) | 0.0038 (11) |
| N1 | 0.0252 (14) | 0.0305 (17) | 0.0200 (13) | 0.0005 (12) | 0.0063 (11) | 0.0030 (11) |
| N2 | 0.0193 (13) | 0.0305 (17) | 0.0179 (13) | 0.0017 (11) | 0.0093 (10) | 0.0011 (11) |
| N3 | 0.0213 (14) | 0.0332 (18) | 0.0226 (14) | −0.0016 (12) | 0.0080 (11) | 0.0057 (12) |
| C1 | 0.0251 (17) | 0.028 (2) | 0.0209 (16) | −0.0040 (14) | 0.0075 (13) | −0.0026 (14) |
| C2 | 0.0274 (18) | 0.034 (2) | 0.0242 (18) | −0.0042 (15) | 0.0092 (14) | −0.0018 (15) |
| C3 | 0.0250 (17) | 0.036 (2) | 0.0281 (17) | 0.0057 (14) | 0.0145 (14) | −0.0007 (15) |
| C4 | 0.0240 (17) | 0.035 (2) | 0.0244 (17) | 0.0043 (14) | 0.0109 (13) | −0.0012 (14) |
| C5 | 0.0239 (17) | 0.041 (2) | 0.0297 (18) | 0.0028 (15) | 0.0147 (14) | 0.0035 (15) |
| C6 | 0.0176 (16) | 0.044 (2) | 0.0348 (19) | −0.0002 (15) | 0.0084 (14) | 0.0032 (16) |
| C7 | 0.0222 (17) | 0.038 (2) | 0.0293 (18) | 0.0033 (15) | 0.0059 (13) | 0.0009 (15) |
| C8 | 0.0256 (17) | 0.031 (2) | 0.0274 (17) | 0.0024 (14) | 0.0093 (14) | 0.0030 (14) |
| C9 | 0.0223 (16) | 0.023 (2) | 0.0285 (17) | 0.0013 (13) | 0.0112 (13) | 0.0003 (14) |
| C10 | 0.0216 (17) | 0.026 (2) | 0.0253 (17) | 0.0004 (13) | 0.0084 (13) | −0.0035 (14) |
| C11 | 0.0197 (16) | 0.027 (2) | 0.0193 (15) | −0.0018 (13) | 0.0082 (12) | −0.0033 (13) |
| C12 | 0.0282 (18) | 0.038 (2) | 0.0221 (16) | 0.0009 (15) | 0.0079 (13) | 0.0041 (15) |
| C13 | 0.0294 (18) | 0.040 (2) | 0.0183 (16) | −0.0014 (15) | 0.0059 (13) | 0.0011 (14) |
| C14 | 0.0256 (17) | 0.036 (2) | 0.0279 (17) | 0.0005 (15) | 0.0063 (14) | 0.0023 (15) |
| C15 | 0.040 (2) | 0.038 (2) | 0.036 (2) | −0.0037 (17) | 0.0099 (16) | −0.0002 (17) |
| C16 | 0.0218 (16) | 0.027 (2) | 0.0207 (16) | 0.0008 (13) | 0.0058 (12) | 0.0031 (13) |
| C17 | 0.0219 (16) | 0.034 (2) | 0.0220 (16) | 0.0008 (14) | 0.0060 (13) | 0.0016 (14) |
| C18 | 0.0276 (17) | 0.037 (2) | 0.0199 (16) | 0.0041 (15) | 0.0078 (13) | 0.0012 (15) |
| C19 | 0.0289 (18) | 0.031 (2) | 0.0299 (18) | 0.0049 (15) | 0.0152 (14) | 0.0062 (15) |
| C20 | 0.0238 (17) | 0.030 (2) | 0.0285 (17) | −0.0012 (14) | 0.0045 (13) | 0.0048 (14) |
| C21 | 0.0266 (17) | 0.029 (2) | 0.0227 (16) | 0.0023 (14) | 0.0091 (13) | 0.0024 (14) |
Geometric parameters (Å, °)
| O1—C1 | 1.241 (4) | C10—C11 | 1.355 (4) |
| O2—C2 | 1.218 (4) | C10—C16 | 1.491 (4) |
| N1—C1 | 1.379 (4) | C12—H12A | 0.990 |
| N1—C2 | 1.396 (4) | C12—H12B | 0.990 |
| N1—C12 | 1.459 (4) | C12—C13 | 1.528 (5) |
| N2—C2 | 1.363 (4) | C13—H13A | 0.990 |
| N2—C3 | 1.452 (4) | C13—H13B | 0.990 |
| N2—C11 | 1.417 (4) | C13—C14 | 1.510 (5) |
| N3—H3N | 0.86 (4) | C14—H14A | 0.990 |
| N3—C9 | 1.413 (4) | C14—H14B | 0.990 |
| N3—C10 | 1.367 (4) | C14—C15 | 1.519 (5) |
| C1—C11 | 1.453 (5) | C15—H15A | 0.980 |
| C3—H3A | 0.990 | C15—H15B | 0.980 |
| C3—H3B | 0.990 | C15—H15C | 0.980 |
| C3—C4 | 1.492 (5) | C16—C17 | 1.390 (4) |
| C4—C5 | 1.396 (5) | C16—C21 | 1.387 (5) |
| C4—C9 | 1.403 (4) | C17—H17 | 0.950 |
| C5—H5 | 0.950 | C17—C18 | 1.386 (4) |
| C5—C6 | 1.379 (5) | C18—H18 | 0.950 |
| C6—H6 | 0.950 | C18—C19 | 1.380 (5) |
| C6—C7 | 1.382 (5) | C19—H19 | 0.950 |
| C7—H7 | 0.950 | C19—C20 | 1.381 (5) |
| C7—C8 | 1.387 (4) | C20—H20 | 0.950 |
| C8—H8 | 0.950 | C20—C21 | 1.392 (4) |
| C8—C9 | 1.383 (5) | C21—H21 | 0.950 |
| C1—N1—C2 | 111.6 (3) | C1—C11—C10 | 128.3 (3) |
| C1—N1—C12 | 124.5 (3) | N1—C12—H12A | 108.9 |
| C2—N1—C12 | 123.8 (2) | N1—C12—H12B | 108.9 |
| C2—N2—C3 | 122.9 (2) | N1—C12—C13 | 113.2 (3) |
| C2—N2—C11 | 111.2 (2) | H12A—C12—H12B | 107.7 |
| C3—N2—C11 | 124.5 (3) | H12A—C12—C13 | 108.9 |
| H3N—N3—C9 | 115 (2) | H12B—C12—C13 | 108.9 |
| H3N—N3—C10 | 115 (2) | C12—C13—H13A | 109.2 |
| C9—N3—C10 | 129.6 (3) | C12—C13—H13B | 109.2 |
| O1—C1—N1 | 122.6 (3) | C12—C13—C14 | 112.2 (3) |
| O1—C1—C11 | 131.4 (3) | H13A—C13—H13B | 107.9 |
| N1—C1—C11 | 105.9 (3) | H13A—C13—C14 | 109.2 |
| O2—C2—N1 | 125.6 (3) | H13B—C13—C14 | 109.2 |
| O2—C2—N2 | 128.5 (3) | C13—C14—H14A | 109.2 |
| N1—C2—N2 | 105.9 (2) | C13—C14—H14B | 109.2 |
| N2—C3—H3A | 108.9 | C13—C14—C15 | 111.9 (3) |
| N2—C3—H3B | 108.9 | H14A—C14—H14B | 107.9 |
| N2—C3—C4 | 113.4 (3) | H14A—C14—C15 | 109.2 |
| H3A—C3—H3B | 107.7 | H14B—C14—C15 | 109.2 |
| H3A—C3—C4 | 108.9 | C14—C15—H15A | 109.5 |
| H3B—C3—C4 | 108.9 | C14—C15—H15B | 109.5 |
| C3—C4—C5 | 120.5 (3) | C14—C15—H15C | 109.5 |
| C3—C4—C9 | 122.2 (3) | H15A—C15—H15B | 109.5 |
| C5—C4—C9 | 117.3 (3) | H15A—C15—H15C | 109.5 |
| C4—C5—H5 | 118.8 | H15B—C15—H15C | 109.5 |
| C4—C5—C6 | 122.5 (3) | C10—C16—C17 | 119.8 (3) |
| H5—C5—C6 | 118.8 | C10—C16—C21 | 121.0 (3) |
| C5—C6—H6 | 120.4 | C17—C16—C21 | 119.1 (3) |
| C5—C6—C7 | 119.2 (3) | C16—C17—H17 | 119.9 |
| H6—C6—C7 | 120.4 | C16—C17—C18 | 120.1 (3) |
| C6—C7—H7 | 120.2 | H17—C17—C18 | 119.9 |
| C6—C7—C8 | 119.7 (3) | C17—C18—H18 | 119.7 |
| H7—C7—C8 | 120.2 | C17—C18—C19 | 120.6 (3) |
| C7—C8—H8 | 119.6 | H18—C18—C19 | 119.7 |
| C7—C8—C9 | 120.9 (3) | C18—C19—H19 | 120.2 |
| H8—C8—C9 | 119.6 | C18—C19—C20 | 119.6 (3) |
| N3—C9—C4 | 122.1 (3) | H19—C19—C20 | 120.2 |
| N3—C9—C8 | 117.7 (3) | C19—C20—H20 | 120.0 |
| C4—C9—C8 | 120.2 (3) | C19—C20—C21 | 120.1 (3) |
| N3—C10—C11 | 126.4 (3) | H20—C20—C21 | 120.0 |
| N3—C10—C16 | 113.5 (3) | C16—C21—C20 | 120.5 (3) |
| C11—C10—C16 | 120.1 (3) | C16—C21—H21 | 119.8 |
| N2—C11—C1 | 105.0 (2) | C20—C21—H21 | 119.8 |
| N2—C11—C10 | 126.6 (3) | ||
| C2—N1—C1—O1 | −176.4 (3) | C9—N3—C10—C16 | 158.5 (3) |
| C2—N1—C1—C11 | 1.5 (4) | N3—C10—C11—N2 | −10.9 (6) |
| C12—N1—C1—O1 | 3.5 (5) | N3—C10—C11—C1 | 165.3 (3) |
| C12—N1—C1—C11 | −178.5 (3) | C16—C10—C11—N2 | 168.2 (3) |
| C3—N2—C2—O2 | 6.5 (5) | C16—C10—C11—C1 | −15.6 (5) |
| C3—N2—C2—N1 | −172.0 (3) | C2—N2—C11—C1 | 5.7 (4) |
| C11—N2—C2—O2 | 173.7 (3) | C2—N2—C11—C10 | −177.3 (3) |
| C11—N2—C2—N1 | −4.8 (4) | C3—N2—C11—C1 | 172.7 (3) |
| C1—N1—C2—O2 | −176.7 (3) | C3—N2—C11—C10 | −10.4 (5) |
| C1—N1—C2—N2 | 2.0 (4) | O1—C1—C11—N2 | 173.5 (3) |
| C12—N1—C2—O2 | 3.4 (5) | O1—C1—C11—C10 | −3.4 (6) |
| C12—N1—C2—N2 | −178.0 (3) | N1—C1—C11—N2 | −4.2 (3) |
| C2—N2—C3—C4 | −136.8 (3) | N1—C1—C11—C10 | 178.9 (3) |
| C11—N2—C3—C4 | 57.7 (4) | C1—N1—C12—C13 | −80.8 (4) |
| N2—C3—C4—C5 | 121.8 (3) | C2—N1—C12—C13 | 99.1 (3) |
| N2—C3—C4—C9 | −61.3 (4) | N1—C12—C13—C14 | −180.0 (2) |
| C3—C4—C5—C6 | 174.0 (3) | C12—C13—C14—C15 | 172.6 (3) |
| C9—C4—C5—C6 | −3.1 (5) | N3—C10—C16—C17 | −52.2 (4) |
| C4—C5—C6—C7 | −1.3 (5) | N3—C10—C16—C21 | 127.0 (3) |
| C5—C6—C7—C8 | 3.1 (5) | C11—C10—C16—C17 | 128.6 (3) |
| C6—C7—C8—C9 | −0.3 (5) | C11—C10—C16—C21 | −52.2 (5) |
| C7—C8—C9—N3 | 174.6 (3) | C10—C16—C17—C18 | 179.6 (3) |
| C7—C8—C9—C4 | −4.3 (5) | C21—C16—C17—C18 | 0.3 (5) |
| C3—C4—C9—N3 | 9.9 (5) | C16—C17—C18—C19 | 0.5 (5) |
| C3—C4—C9—C8 | −171.2 (3) | C17—C18—C19—C20 | −0.6 (5) |
| C5—C4—C9—N3 | −173.1 (3) | C18—C19—C20—C21 | −0.2 (5) |
| C5—C4—C9—C8 | 5.8 (5) | C10—C16—C21—C20 | 179.7 (3) |
| C10—N3—C9—C4 | 37.7 (5) | C17—C16—C21—C20 | −1.1 (5) |
| C10—N3—C9—C8 | −141.3 (3) | C19—C20—C21—C16 | 1.1 (5) |
| C9—N3—C10—C11 | −22.3 (6) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H3N···O1i | 0.86 (4) | 2.10 (4) | 2.944 (3) | 165 (3) |
| C8—H8···O1i | 0.95 | 2.57 | 3.326 (4) | 136 |
Symmetry codes: (i) −x+3/2, y+1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BH2272).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Hulme, C. & Gore, V. (2003). Curr. Med. Chem.10, 51–80. [DOI] [PubMed]
- Hulme, C., Ma, L., Romano, J. J., Morton, G., Tang, S.-Y., Cherrier, M.-P., Choi, S., Salvino, J. & Labaudiniere, R. (2000). Tetrahedron Lett.41, 1889–1893.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst.41, 466–470.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810005477/bh2272sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810005477/bh2272Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report