Abstract
The structure of the the title compound, [Cu2(C7H3ClO3)2(C12H8N2)2]·2H2O, consists of a dimeric unit involving a planar Cu2O2 group arranged around an inversion center. The coordination sphere of the CuII atom can be described as an elongated distorted square pyramid where the basal plane is formed by the two N atoms of the 1,10-phenanthroline molecule and the two O atoms of the hydroxychlorobenzoate (hcbe) anion. The long apical Cu—O distance of 2.569 (2) Å involves the O atom of a symmetry-related hcbe anion, building up the dinuclear unit. Each dinuclear unit is connected through O—H⋯O hydrogen bonds involving two water molecules, resulting in an R
4
2(8) graph-set motif and building up an infinite chain parallel to (10 ). C—H⋯O interactions further stabilize the chain.
). C—H⋯O interactions further stabilize the chain.
Related literature
For our ongoing investigation of the nature of π–π stacking, see: Su & Xu (2004 ▶); Xu et al. (2007 ▶). For related structures, see: Yang et al. (2006 ▶); Garland et al. (1987 ▶); Li et al. (1995 ▶); Fan & Zhu (2005 ▶); Song et al. (2007 ▶). For a structural discussion on hydrogen bonding, see: Etter et al. (1990 ▶); Bernstein et al. (1995 ▶).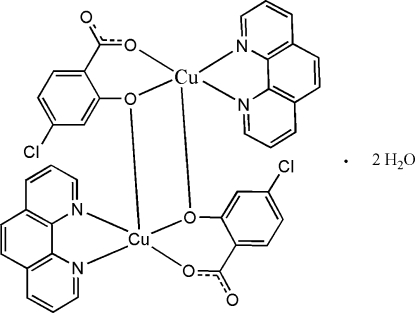
Experimental
Crystal data
[Cu2(C7H3ClO3)2(C12H8N2)2]·2H2O
M r = 864.60
Monoclinic,
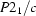
a = 8.1941 (17) Å
b = 18.851 (4) Å
c = 11.873 (3) Å
β = 105.993 (8)°
V = 1763.0 (6) Å3
Z = 2
Mo Kα radiation
μ = 1.42 mm−1
T = 294 K
0.33 × 0.30 × 0.22 mm
Data collection
Rigaku R-AXIS RAPID IP diffractometer
Absorption correction: multi-scan (ABSCOR; Higashi, 1995 ▶) T min = 0.656, T max = 0.730
18894 measured reflections
3163 independent reflections
2162 reflections with I > 2σ(I)
R int = 0.047
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.103
S = 1.03
3163 reflections
244 parameters
H-atom parameters constrained
Δρmax = 0.58 e Å−3
Δρmin = −0.36 e Å−3
Data collection: PROCESS-AUTO (Rigaku, 1998 ▶); cell refinement: PROCESS-AUTO; data reduction: CrystalStructure (Rigaku/MSC, 2002 ▶); program(s) used to solve structure: SIR92 (Altomare et al., 1993 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810008354/dn2542sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810008354/dn2542Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1W—H1A⋯O2 | 0.90 | 1.92 | 2.817 (4) | 175 |
| O1W—H1B⋯O2i | 0.88 | 2.13 | 2.921 (4) | 150 |
| C10—H10⋯O2ii | 0.93 | 2.42 | 3.277 (5) | 153 |
| C17—H17⋯O1Wi | 0.93 | 2.58 | 3.487 (4) | 166 |
Symmetry codes: (i) 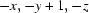 ; (ii)
; (ii) 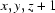 .
.
Acknowledgments
The work was supported by the ZIJIN project of Zhejiang University, China.
supplementary crystallographic information
Comment
As part of our ongoing investigation on the nature of π-π stacking (Su & Xu, 2004; Xu et al., 2007), the title CuII compound incorporating 2-hydroxy-4-chlorobenzoate (hcbe) ligand has recently been prepared in the laboratory and its crystal structure is reported here.
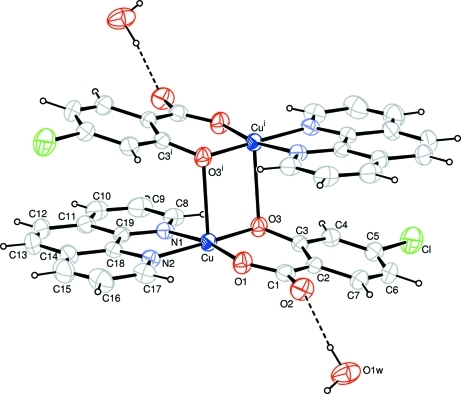
The structure of the the title compound, (C19H11ClCuN2O3).(H2O), consists of a dimeric unit involving a planar Cu2O2 group arranged around inversion center The coordination sphere of the CuII can be described as an elongated distorted square pyramid where the basal plane is formed by the two N atoms of the 1,10-phenanthroline molecule and the two O atoms of the hydroxychlorobenzoate (hcbe) anion. The long apical Cu-O3 distance of 2.569 (2)A involves the O3 atom of the symmetry related hcbe anion [symmetry code (i) i-x,1-y,1-z] building up the dinuclear unit (Fig. 1).
This apical Cu-O3 distance is 0.674 (3) Å longer than Cu—O3 bond distance in the basal coordination plane, showing the Jahn-Teller distorted square-pyramidal coordination geometry around the CuII cation. A patially overlapped arrangement is observed between the nearly parallel C2-C7 phenyl ring and C11-C19 phen ring system [dihedral angle 14.25°]. The centroid to centroid distance is 3.649 (3) Å and the perpendicular distance of the centroid to the rings is 3.456 and 3.571 Å respectively suggesting a weak π-π stacking comparable to that found in the related CuII complexes (Garland et al., 1987; Li et al., 1995; Fan & Zhu, 2005; Song et al., 2007) and also to the NiII complex of 2,4-dihydroxybenzoate (Yang et al., 2006).
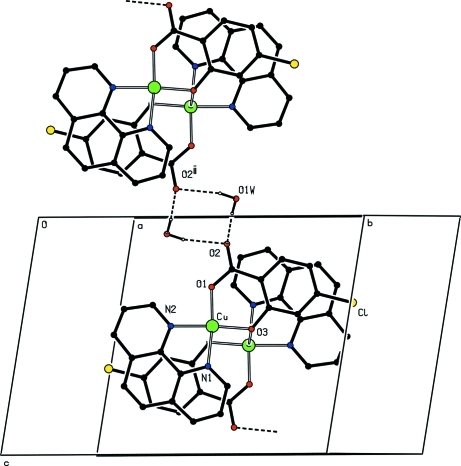
Each dinuclear unit are connected through O-H···O hydrogen bonds involving two water molecules resulting in a R24(8) graph set motif ( Etter et al., 1990, Bernstein et al., 1995) and building up an infinite chain parallel to the (1 0 -1) plane. C-H···O interactions further stabilize the chain. (Table 1; Fig. 2).
Experimental
An ethanol-water solution (20 ml, 1:3) containing 2-hydroxy-4-chlorobenzoic acid (0.173 g, 1 mmol), Na2CO3 (0.053 g, 0.5 mmol) and CuCl2.2H2O (0.085 g, 0.5 mmol) was refluxed for 6 h, then phenanthroline hydrate (0.99 g, 1 mmol) was added into the solution and the mixture was refluxed for further 0.5 h. After cooling to room temperature the solution was filtered. Single crystals of the title compound were obtained from the filtrate after one week.
Refinement
Water H atoms were located in a difference Fourier map and refined as riding in as-found relative positions with Uiso(H) = 1.5Ueq(O). Other H atoms were placed in calculated positions with C—H = 0.93 Å and refined in riding mode with Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
The dinuclear molecular structure of the title compound with 30% probability displacement (arbitrary spheres for H atoms) [symmetry code: (i) 1-x, 1-y, 1-z].
Fig. 2.
Partial packing view showing the formation of the chain through the O-H···O hydrogen bonds. H atoms not involved in hydrogen bondings have been omitted for clarity. H bonds are shown as dashed line.[Symmetry codes: (ii) -x, -y+1, -z]
Crystal data
| [Cu2(C7H3ClO3)2(C12H8N2)2]·2H2O | F(000) = 876 |
| Mr = 864.60 | Dx = 1.629 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 5352 reflections |
| a = 8.1941 (17) Å | θ = 2.2–24.2° |
| b = 18.851 (4) Å | µ = 1.42 mm−1 |
| c = 11.873 (3) Å | T = 294 K |
| β = 105.993 (8)° | Prism, blue |
| V = 1763.0 (6) Å3 | 0.33 × 0.30 × 0.22 mm |
| Z = 2 |
Data collection
| Rigaku R-AXIS RAPID IP diffractometer | 3163 independent reflections |
| Radiation source: fine-focus sealed tube | 2162 reflections with I > 2σ(I) |
| graphite | Rint = 0.047 |
| Detector resolution: 10.0 pixels mm-1 | θmax = 25.2°, θmin = 2.1° |
| ω scans | h = −9→9 |
| Absorption correction: multi-scan (ABSCOR; Higashi, 1995) | k = −22→22 |
| Tmin = 0.656, Tmax = 0.730 | l = −13→14 |
| 18894 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.103 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0509P)2 + 0.5996P] where P = (Fo2 + 2Fc2)/3 |
| 3163 reflections | (Δ/σ)max = 0.001 |
| 244 parameters | Δρmax = 0.58 e Å−3 |
| 0 restraints | Δρmin = −0.35 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cu | 0.28977 (4) | 0.501289 (19) | 0.46012 (3) | 0.04370 (15) | |
| Cl | 0.68680 (17) | 0.80306 (6) | 0.36270 (13) | 0.1107 (5) | |
| N1 | 0.2931 (3) | 0.50654 (13) | 0.6296 (2) | 0.0456 (6) | |
| N2 | 0.1240 (3) | 0.42264 (13) | 0.4601 (2) | 0.0480 (6) | |
| O1 | 0.2426 (3) | 0.49404 (12) | 0.2962 (2) | 0.0596 (6) | |
| O2 | 0.2234 (3) | 0.52593 (16) | 0.1153 (2) | 0.0750 (7) | |
| O3 | 0.4451 (3) | 0.57734 (11) | 0.47282 (18) | 0.0507 (5) | |
| C1 | 0.2743 (4) | 0.5384 (2) | 0.2219 (3) | 0.0555 (9) | |
| C2 | 0.3717 (4) | 0.60392 (18) | 0.2646 (3) | 0.0512 (8) | |
| C3 | 0.4487 (4) | 0.61966 (16) | 0.3838 (3) | 0.0472 (7) | |
| C4 | 0.5430 (4) | 0.68293 (17) | 0.4110 (3) | 0.0577 (9) | |
| H4 | 0.5950 | 0.6941 | 0.4889 | 0.069* | |
| C5 | 0.5593 (5) | 0.72807 (19) | 0.3250 (4) | 0.0718 (11) | |
| C6 | 0.4837 (5) | 0.7141 (2) | 0.2088 (4) | 0.0851 (14) | |
| H6 | 0.4947 | 0.7455 | 0.1509 | 0.102* | |
| C7 | 0.3919 (5) | 0.6528 (2) | 0.1802 (3) | 0.0711 (11) | |
| H7 | 0.3408 | 0.6431 | 0.1015 | 0.085* | |
| C8 | 0.3797 (4) | 0.55026 (19) | 0.7127 (3) | 0.0569 (9) | |
| H8 | 0.4424 | 0.5868 | 0.6923 | 0.068* | |
| C9 | 0.3792 (5) | 0.5429 (2) | 0.8297 (3) | 0.0728 (11) | |
| H9 | 0.4401 | 0.5745 | 0.8857 | 0.087* | |
| C10 | 0.2897 (5) | 0.4896 (2) | 0.8616 (4) | 0.0765 (13) | |
| H10 | 0.2892 | 0.4847 | 0.9394 | 0.092* | |
| C11 | 0.1979 (5) | 0.4418 (2) | 0.7767 (3) | 0.0635 (10) | |
| C12 | 0.1001 (6) | 0.3835 (3) | 0.7997 (4) | 0.0854 (14) | |
| H12 | 0.0971 | 0.3746 | 0.8761 | 0.102* | |
| C13 | 0.0125 (6) | 0.3412 (3) | 0.7126 (5) | 0.0876 (14) | |
| H13 | −0.0510 | 0.3041 | 0.7306 | 0.105* | |
| C14 | 0.0135 (5) | 0.3514 (2) | 0.5930 (4) | 0.0675 (11) | |
| C15 | −0.0751 (6) | 0.3107 (2) | 0.4968 (5) | 0.0882 (14) | |
| H15 | −0.1426 | 0.2731 | 0.5076 | 0.106* | |
| C16 | −0.0626 (5) | 0.3259 (2) | 0.3884 (5) | 0.0849 (13) | |
| H16 | −0.1223 | 0.2990 | 0.3247 | 0.102* | |
| C17 | 0.0394 (5) | 0.38181 (19) | 0.3716 (3) | 0.0639 (10) | |
| H17 | 0.0486 | 0.3909 | 0.2966 | 0.077* | |
| C18 | 0.1106 (4) | 0.40810 (17) | 0.5688 (3) | 0.0505 (8) | |
| C19 | 0.2019 (4) | 0.45334 (17) | 0.6605 (3) | 0.0499 (8) | |
| O1W | 0.0002 (4) | 0.59767 (16) | −0.0734 (2) | 0.0934 (9) | |
| H1A | 0.0653 | 0.5735 | −0.0124 | 0.140* | |
| H1B | −0.0941 | 0.5733 | −0.0960 | 0.140* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu | 0.0459 (2) | 0.0543 (2) | 0.0309 (2) | −0.00153 (17) | 0.01078 (16) | 0.00139 (17) |
| Cl | 0.1106 (10) | 0.0650 (6) | 0.1553 (13) | −0.0153 (6) | 0.0347 (9) | 0.0283 (7) |
| N1 | 0.0459 (14) | 0.0589 (15) | 0.0335 (14) | 0.0089 (13) | 0.0134 (11) | −0.0016 (13) |
| N2 | 0.0424 (15) | 0.0544 (15) | 0.0472 (17) | 0.0028 (12) | 0.0124 (12) | −0.0025 (13) |
| O1 | 0.0688 (16) | 0.0716 (15) | 0.0349 (13) | −0.0104 (12) | 0.0085 (11) | −0.0014 (11) |
| O2 | 0.0772 (18) | 0.117 (2) | 0.0277 (14) | 0.0076 (16) | 0.0085 (12) | 0.0011 (13) |
| O3 | 0.0577 (13) | 0.0586 (13) | 0.0357 (12) | −0.0060 (10) | 0.0125 (10) | 0.0031 (10) |
| C1 | 0.0470 (19) | 0.081 (2) | 0.038 (2) | 0.0107 (18) | 0.0103 (15) | 0.0060 (18) |
| C2 | 0.0443 (18) | 0.069 (2) | 0.0419 (19) | 0.0106 (16) | 0.0144 (15) | 0.0163 (16) |
| C3 | 0.0443 (18) | 0.0539 (18) | 0.046 (2) | 0.0079 (15) | 0.0176 (15) | 0.0062 (16) |
| C4 | 0.057 (2) | 0.0537 (19) | 0.064 (2) | 0.0052 (16) | 0.0197 (18) | 0.0051 (17) |
| C5 | 0.060 (2) | 0.058 (2) | 0.099 (3) | 0.0067 (18) | 0.025 (2) | 0.025 (2) |
| C6 | 0.075 (3) | 0.095 (3) | 0.089 (3) | 0.008 (2) | 0.028 (3) | 0.053 (3) |
| C7 | 0.063 (2) | 0.099 (3) | 0.052 (2) | 0.010 (2) | 0.0164 (19) | 0.028 (2) |
| C8 | 0.056 (2) | 0.071 (2) | 0.042 (2) | 0.0117 (17) | 0.0109 (16) | −0.0050 (17) |
| C9 | 0.074 (3) | 0.102 (3) | 0.038 (2) | 0.021 (2) | 0.0075 (19) | −0.011 (2) |
| C10 | 0.078 (3) | 0.119 (4) | 0.040 (2) | 0.034 (3) | 0.028 (2) | 0.013 (2) |
| C11 | 0.058 (2) | 0.089 (3) | 0.052 (2) | 0.024 (2) | 0.0296 (18) | 0.018 (2) |
| C12 | 0.076 (3) | 0.118 (4) | 0.078 (3) | 0.031 (3) | 0.050 (3) | 0.045 (3) |
| C13 | 0.074 (3) | 0.089 (3) | 0.117 (4) | 0.009 (2) | 0.055 (3) | 0.040 (3) |
| C14 | 0.054 (2) | 0.064 (2) | 0.094 (3) | 0.0047 (18) | 0.035 (2) | 0.016 (2) |
| C15 | 0.077 (3) | 0.062 (2) | 0.129 (5) | −0.013 (2) | 0.034 (3) | 0.004 (3) |
| C16 | 0.069 (3) | 0.069 (3) | 0.113 (4) | −0.014 (2) | 0.019 (3) | −0.024 (3) |
| C17 | 0.057 (2) | 0.062 (2) | 0.069 (3) | 0.0008 (18) | 0.0128 (19) | −0.0129 (19) |
| C18 | 0.0417 (18) | 0.0575 (19) | 0.055 (2) | 0.0093 (15) | 0.0183 (16) | 0.0081 (17) |
| C19 | 0.0462 (19) | 0.062 (2) | 0.047 (2) | 0.0175 (16) | 0.0226 (15) | 0.0131 (16) |
| O1W | 0.097 (2) | 0.111 (2) | 0.0617 (18) | 0.0296 (18) | 0.0041 (15) | 0.0005 (16) |
Geometric parameters (Å, °)
| Cu—O1 | 1.882 (2) | C8—C9 | 1.397 (5) |
| Cu—O3 | 1.895 (2) | C8—H8 | 0.9300 |
| Cu—O3i | 2.569 (2) | C9—C10 | 1.358 (6) |
| Cu—N1 | 2.007 (3) | C9—H9 | 0.9300 |
| Cu—N2 | 2.011 (3) | C10—C11 | 1.405 (6) |
| Cl—C5 | 1.741 (4) | C10—H10 | 0.9300 |
| N1—C8 | 1.331 (4) | C11—C19 | 1.406 (4) |
| N1—C19 | 1.360 (4) | C11—C12 | 1.430 (6) |
| N2—C17 | 1.332 (4) | C12—C13 | 1.346 (6) |
| N2—C18 | 1.353 (4) | C12—H12 | 0.9300 |
| O1—C1 | 1.292 (4) | C13—C14 | 1.435 (6) |
| O2—C1 | 1.241 (4) | C13—H13 | 0.9300 |
| O3—C3 | 1.331 (4) | C14—C15 | 1.401 (6) |
| C1—C2 | 1.482 (5) | C14—C18 | 1.409 (5) |
| C2—C7 | 1.404 (4) | C15—C16 | 1.350 (6) |
| C2—C3 | 1.413 (4) | C15—H15 | 0.9300 |
| C3—C4 | 1.409 (5) | C16—C17 | 1.393 (5) |
| C4—C5 | 1.363 (5) | C16—H16 | 0.9300 |
| C4—H4 | 0.9300 | C17—H17 | 0.9300 |
| C5—C6 | 1.374 (6) | C18—C19 | 1.423 (5) |
| C6—C7 | 1.370 (6) | O1W—H1A | 0.8969 |
| C6—H6 | 0.9300 | O1W—H1B | 0.8747 |
| C7—H7 | 0.9300 | ||
| O1—Cu—O3 | 94.54 (9) | C2—C7—H7 | 118.6 |
| O1—Cu—N1 | 169.27 (10) | N1—C8—C9 | 121.9 (4) |
| O3—Cu—N1 | 93.42 (10) | N1—C8—H8 | 119.1 |
| O1—Cu—N2 | 90.01 (10) | C9—C8—H8 | 119.1 |
| O3—Cu—N2 | 175.25 (9) | C10—C9—C8 | 120.1 (4) |
| N1—Cu—N2 | 81.90 (11) | C10—C9—H9 | 120.0 |
| O1—Cu—O3i | 101.17 (9) | C8—C9—H9 | 120.0 |
| O3—Cu—O3i | 85.40 (9) | C9—C10—C11 | 119.8 (4) |
| N1—Cu—O3i | 86.62 (8) | C9—C10—H10 | 120.1 |
| N2—Cu—O3i | 95.07 (9) | C11—C10—H10 | 120.1 |
| C8—N1—C19 | 118.5 (3) | C10—C11—C19 | 116.9 (4) |
| C8—N1—Cu | 129.1 (2) | C10—C11—C12 | 125.0 (4) |
| C19—N1—Cu | 112.2 (2) | C19—C11—C12 | 118.1 (4) |
| C17—N2—C18 | 118.3 (3) | C13—C12—C11 | 121.2 (4) |
| C17—N2—Cu | 129.1 (3) | C13—C12—H12 | 119.4 |
| C18—N2—Cu | 112.4 (2) | C11—C12—H12 | 119.4 |
| C1—O1—Cu | 129.5 (2) | C12—C13—C14 | 122.2 (4) |
| C3—O3—Cu | 123.5 (2) | C12—C13—H13 | 118.9 |
| O2—C1—O1 | 119.9 (3) | C14—C13—H13 | 118.9 |
| O2—C1—C2 | 120.3 (3) | C15—C14—C18 | 116.3 (4) |
| O1—C1—C2 | 119.8 (3) | C15—C14—C13 | 126.2 (4) |
| C7—C2—C3 | 118.0 (3) | C18—C14—C13 | 117.5 (4) |
| C7—C2—C1 | 117.4 (3) | C16—C15—C14 | 120.2 (4) |
| C3—C2—C1 | 124.6 (3) | C16—C15—H15 | 119.9 |
| O3—C3—C4 | 117.2 (3) | C14—C15—H15 | 119.9 |
| O3—C3—C2 | 124.6 (3) | C15—C16—C17 | 120.3 (4) |
| C4—C3—C2 | 118.2 (3) | C15—C16—H16 | 119.9 |
| C5—C4—C3 | 121.2 (4) | C17—C16—H16 | 119.9 |
| C5—C4—H4 | 119.4 | N2—C17—C16 | 121.8 (4) |
| C3—C4—H4 | 119.4 | N2—C17—H17 | 119.1 |
| C6—C5—C4 | 121.5 (4) | C16—C17—H17 | 119.1 |
| C6—C5—Cl | 119.1 (3) | N2—C18—C14 | 123.2 (3) |
| C4—C5—Cl | 119.4 (3) | N2—C18—C19 | 116.4 (3) |
| C5—C6—C7 | 118.5 (3) | C14—C18—C19 | 120.4 (3) |
| C5—C6—H6 | 120.8 | N1—C19—C11 | 122.8 (3) |
| C7—C6—H6 | 120.8 | N1—C19—C18 | 116.6 (3) |
| C6—C7—C2 | 122.7 (4) | C11—C19—C18 | 120.6 (3) |
| C6—C7—H7 | 118.6 | H1A—O1W—H1B | 105.1 |
Symmetry codes: (i) −x+1, −y+1, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H1A···O2 | 0.90 | 1.92 | 2.817 (4) | 175 |
| O1W—H1B···O2ii | 0.88 | 2.13 | 2.921 (4) | 150 |
| C10—H10···O2iii | 0.93 | 2.42 | 3.277 (5) | 153 |
| C17—H17···O1Wii | 0.93 | 2.58 | 3.487 (4) | 166 |
Symmetry codes: (ii) −x, −y+1, −z; (iii) x, y, z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: DN2542).
References
- Altomare, A., Cascarano, G., Giacovazzo, C. & Guagliardi, A. (1993). J. Appl. Cryst.26, 343–350.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Etter, M. C., MacDonald, J. C. & Bernstein, J. (1990). Acta Cryst. B46, 256–262. [DOI] [PubMed]
- Fan, S.-R. & Zhu, L.-G. (2005). Chin. J. Chem 23, 1292–1296.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Garland, M. T., Grandjean, D. & Spodine, E. (1987). Acta Cryst. C43, 1910–1912.
- Higashi, T. (1995). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Li, M., Zou, J.-Z., Xu, Z., You, X.-Z. & Huang, X.-Y. (1995). Polyhedron, 14, 639–644.
- Rigaku (1998). PROCESS-AUTO Rigaku Corporation, Tokyo, Japan.
- Rigaku/MSC (2002). CrystalStructure MSC, The Woodlands, Texas, USA, and Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Song, J.-F., Chen, Y., Li, Z.-G., Zhou, R.-S., Xu, X.-Y., Xu, J.-Q. & Wang, T.-G. (2007). Polyhedron, 26, 4397–4402
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Su, J.-R. & Xu, D.-J. (2004). J. Coord. Chem.57, 223–229.
- Xu, D.-J., Zhang, B.-Y., Su, J.-R. & Nie, J.-J. (2007). Acta Cryst. C63, m622–m624. [DOI] [PubMed]
- Yang, Q., Zhang, L. & Xu, D.-J. (2006). Acta Cryst. E62, m2678–m2680.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810008354/dn2542sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810008354/dn2542Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report