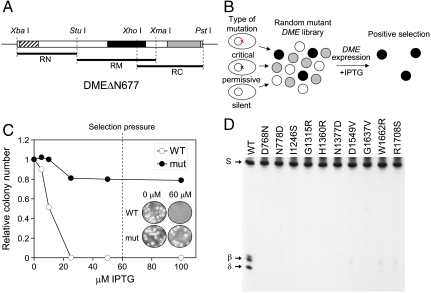
Fig. 2.
Isolation of mutant DME proteins sensitive to random amino acid change. (A) DMEΔN677 was divided into three pieces (RN, RM, and RC) and subjected to random mutagenesis by error-prone PCR. Three conserved domains—domain A, glycosylase domain, and domain B—are indicated by hatched, solid, and shaded boxes, respectively. The restriction sites used for cloning are indicated above the DME fragment. (B) Screening scheme of a random mutant library. A mutant library was generated and screened for viability in the presence of IPTG to induce DME expression. Only cells with a critical mutation(s) will survive due to a loss of 5mC glycosylase activity that would otherwise produce critical damages in E. coli. Surviving colonies were positively selected and analyzed for enzyme activity. (C) The cells expressing catalytically inactive DME survived, whereas the cells expressing WT DME died as IPTG concentrations increased. A dashed line represents the concentration of IPTG (60 μM) in media that exerted selection pressure. (Inset) Colony formation of wild-type and mutant (D1304N) DME transformants at 0 and 60 μM IPTG. (D) 5mC glycosylase activity of some representative mutant DME proteins. WT and mutant proteins were reacted with methylated oligonucleotide substrate and separated on a denaturing polyacrylamide gel.