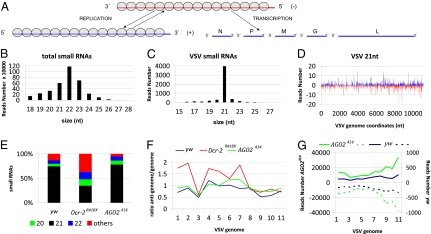
Fig. 3.
Virus-derived siRNAs in VSV infected flies. (A) Structure and organization of the VSV genome. The 11-kb long negative-strand RNA is the template for synthesis of five monocistronic mRNAs that encode the viral proteins N, P, M, G, and L. During replication, the negative-strand is the template for the synthesis of the full-length positive-strand RNA (antigenome), which will in turn serve as a template for the production of the genomic RNA. The N protein is tightly associated with the genome and antigenome RNA (gray circles). (B) Size distribution of total small RNAs in VSV-infected yw flies. (C) Size distribution of small RNAs aligned against the viral genome in VSV-infected yw flies. (D) Profile of 21 nt VSV-derived reads along the VSV genome. To comply with the orientation of the VSV reference sequence (NC_001560) used for all of the analyses, the genome is represented under its antigenomic polarity. Each VSV-derived small RNAs (vsiRNA) is represented by the position of its first nucleotide. The vsiRNAs matching the antigenome and the genome are shown in blue and red, respectively. The horizontal axis represents the antigenome coordinates. (E) Relative abundance of VSV-derived small RNAs of 20, 21, 22 nt and other sizes (19, 23–28 nt) in yw, Dcr-2R416X, and AGO2414 flies. (F) Relative strand representation of the 21 nt reads expressed as the ratio between the number of reads matching the antigenome over the number of reads matching the genome using 1,000 bp contiguous windows (1–11). (G) Representation of VSV-derived vsiRNAs from yw (blue) and AGO2414 (green) flies expressed as the sum of the reads over 1,000-bp contiguous windows covering the whole genome. The vsiRNAs matching the antigenome and genome are showed as solid and dotted lines, respectively. The sum of the reads corresponding to each window is represented in the vertical axis.