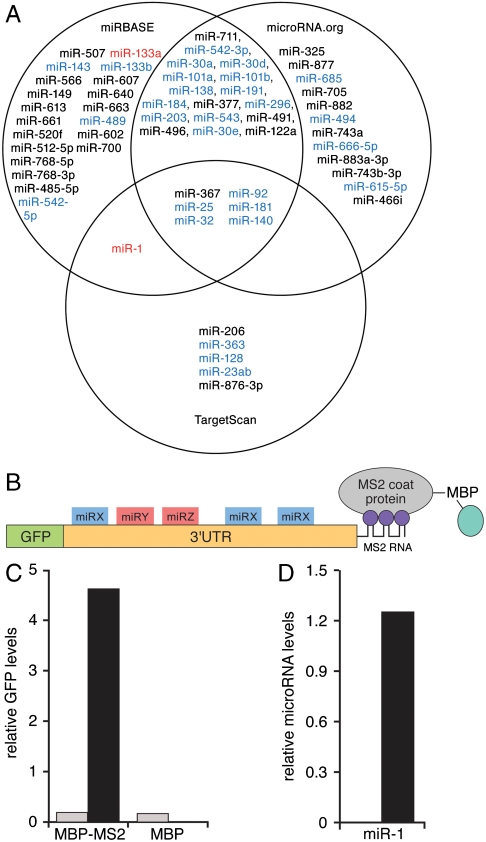
Fig. 1.
Characterization of Hand2-associated microRNAs. (A) Venn diagram of predicted Hand2 3′UTR-associated microRNAs from three algorithms [miRBase (now MicroCosm), TargetScan, and microRNA.org]. MicroRNAs expressed in heart are shown in blue. miR-1 and miR-133a, identified in the affinity purification assay (see below) are shown in red. MicroRNAs predicted to regulate Hand2 but not expressed in heart are shown in black. (B) Schematic of the MS2 affinity purification assay. An MS2-tagged Hand2 3′UTR is expressed downstream of a GFP reporter. Purification of the transcript is mediated by the affinity of the MS2 coat protein (expressed as a fusion with maltose binding protein, MBP) for the MS2 RNA sequence tag. (C) Real-time PCR quantification of GFP-Hand2 3′UTR reporter transcripts expressed in HEK293 cells and purified on an MBP-MS2 affinity column. MBP represents a column lacking the MS2 binding protein. MS2-tagged and untagged transcripts are shown in black and gray, respectively. (D) Taqman PCR analysis of miR-1 associated with the Hand2 3′UTR expressed in differentiated C2C12 cells and purified on an MBP-MS2 affinity column. No binding was detected when the Hand2 3′UTR was inserted in reverse orientation.