Abstract
Effective methods to detect and quantify functionally linked regulatory proteins in complex biological samples are essential for investigating mammalian signaling pathways. Traditional immunoassays depend on proprietary reagents that are difficult to generate and multiplex, whereas global proteomic profiling can be tedious and can miss low abundance proteins. Here, we report a target-driven liquid chromatography-tandem mass spectrometry (LC-MS/MS) strategy for selectively examining the levels of multiple low abundance components of signaling pathways which are refractory to standard shotgun screening procedures and hence appear limited in current MS/MS repositories. Our stepwise approach consists of: (i) synthesizing microscale peptide arrays, including heavy isotope-labeled internal standards, for use as high quality references to (ii) build empirically validated high density LC-MS/MS detection assays with a retention time scheduling system that can be used to (iii) identify and quantify endogenous low abundance protein targets in complex biological mixtures with high accuracy by correlation to a spectral database using new software tools. The method offers a flexible, rapid, and cost-effective means for routine proteomic exploration of biological systems including “label-free” quantification, while minimizing spurious interferences. As proof-of-concept, we have examined the abundance of transcription factors and protein kinases mediating pluripotency and self-renewal in embryonic stem cell populations.
Biological processes are controlled by signaling pathways and co-expression networks. Monitoring the expression levels of critical, but often low abundance, regulatory factors is therefore essential for mechanistic understanding of cellular function (1–3). LC-MS/MS1 is an increasingly popular technique for characterizing biological samples. In a typical “shotgun” proteomics study, a protein mixture is proteolytically digested and the resulting peptides separated by nanoflow LC prior to ionization and fragmentation in the gas phase (4). The recorded MS/MS spectra are subsequently matched to known protein sequences using a protein sequence database search algorithm (5, 6). Despite the capability of modern instrumentation to resolve thousands of peptides in a single analysis, the extreme complexity and dynamic range imbalance of mammalian proteomes pose unresolved challenges. In particular, low abundance proteins are often missed due to undersampling (4) and difficulties in interpreting noisy MS/MS spectra (7). Innovative new methods are therefore needed to detect and quantify the components of signaling systems across different samples reliably and consistently.
Target-driven LC-MS/MS procedures in which only preselected precursor ions corresponding to targets of interest are subject to fragmentation can markedly enhance limits of detection and quantitation (8–11). In proteomics, multiple reaction monitoring (MRM; also called selected reaction monitoring), which involves two consecutive stages of MS filtering on continuous ion beam instruments, is increasingly popular as a means of measuring the abundance of multiple targets (10). Similarly, precursor ions can be continuously isolated and fragmented using trap-type instruments (i.e. pseudo-MRM), termed targeted peptide monitoring (TPM) (10) by us and peptide ion monitoring (PIM) by others (8). Prior knowledge of peptide detectability, chromatographic retention characteristics, and MS/MS peak intensity patterns can be used to confirm protein identities (12, 13).
Although in principle, targeted LC-MS/MS assays can potentially be generated for any protein of interest (14, 15), significant practical hurdles must be overcome. First, a robust assay development pipeline must be devised. Second, target feature selection and experimental parameters must be geared toward achieving the highest possible detection sensitivity and specificity despite the underrepresentation of low abundance (e.g. signaling) proteins in current proteomic repositories. Finally, rigorous quality controls are needed to contend with the contextual complexity of biological specimens that exhibit confounding “interferences” to confirm tentative molecular identities and abundance estimates.
To this end, we report a versatile strategy for rapidly generating multiplexed LC-MS/MS assays that can be used to detect and quantify reliably the expression of low abundance components of signaling pathways in a single experiment, including signaling proteins never detected by mass spectrometry before. The method combines microscale SPOT membrane synthesis (16–20) of reference peptide arrays, empirical assay optimization and scheduling using chromatographic markers to select precisely the most informative and sensitive product ion features combinations for these targets in a given sample background, and application of spectral scoring that can increase detection sensitivity by up to 2 orders of magnitude relative to traditional MS/MS scan interpretation. We apply our method to develop customized assays for tracking low abundance, typically difficult to detect regulatory components of core pluripotency transcriptional network in nuclear extracts from JAK/STAT signaling activated mouse embryonic stem cells (mESCs) (21).
EXPERIMENTAL PROCEDURES
Target Selection and SPOT Membrane Peptide Synthesis
Fully tryptic peptide sequences suitable for the SPOT synthesis were predicted by in silico digestion and basic selection criteria: candidates were allowed for one missed tryptic cleavage site but excluding reactive cysteine and methionine residues and histidine where possible to avoid, e.g. higher charge states during electrospray ionization. Also, the length of peptides was limited to 9–15 amino acids to assure reasonable yield in peptide synthesis. Moreover, an SVM algorithm trained on precursor ion intensities from ∼1,000 identified peptide sequences was used to predict which sequence is likely to be detected in the mass spectrometer and was used to rank peptides without applying any cutoff. In addition, identical human peptides were listed, wherever possible. Then 2–15 top ranking peptides, depending on availability and matching all of the above criteria, were synthesized using microscale Fmoc chemistry on a derivatized cellulose membrane using a fully automated commercial peptide synthesizer (JPT Peptide Technologies, Berlin, Germany). To introduce stable isotope labels, heavy lysine (K) or arginine (R) containing 13C and 15N atoms (enriched to >97%; JPT Peptide Technologies) were incorporated at the C-terminal tryptic residue followed by a cleavable 3-amino acid long “universal” quantitation tag (supplemental Fig. 1B), e.g. 20 ng of peptide was then treated with 2 ng of trypsin in 50 mm ammonium bicarbonate, pH 7.8, for 3 h at 37 °C to cleave off the peptide tag. After stopping the reaction with 1% formic acid (final concentration) and 5× dilution, the peptide was injected into the Orbitrap mass spectrometer using syringe pump with a flow rate of 1 μl/min. Relative abundance from the tag (m/z 276.1554) to the target peptide precursor was calculated from Orbitrap precursor mass spectrum with resolution R = 60,000 to obtain an abundance factor. Also, for the purpose of absolute quantification of the precursor ion intensity, a high purity (>97%) copy of the SLG tag was synthesized to establish an external standard curve. For all other (unlabeled or labeled) peptides, individual spots were cut out after synthesis and the peptides solubilized off the membrane using aqueous ammonia solution and analyzed using an ion trap, Orbitrap, or triple quadrupole instrument in a targeted MS/MS mode (i.e. TPM or MRM). About 50 nmol is synthesized per spot (22), typically with a purity of 80–90%, although yield varies with the target sequence. This degree of purity is enough to establish high quality MS/MS spectra for a reference library, for assay design and optimization, and for use as internal standards in spike-in assays.
Mouse and Yeast Sample Preparation
Parental mESCs (E14Tg2a line) were cultured in 15% FBS (900-108; Gemini Biological Products, West Sacramento, CA) supplemented with 500 pm LIF (mLIF; ESG11-7; Chemicon, Temecula, CA) using gelatin-coated tissue culture flasks, as described previously(21). To initiate differentiation, LIF was withheld for 48 h before harvesting. For all studies, 15% knockout serum (KnockoutTM 10828-028; Invitrogen, Carlsbad, CA) was used. A total of 6 × 107 cells (∼100-μl volume) were harvested, washed twice with PBS, flash frozen, and stored at −80 °C. The cells were thawed on ice by the addition of 500 μl of 1× lysis buffer (10× LB = 100 mm Hepes, pH 7.9, 15 mm MgCl2, 100 mm KCl) with 5 μl of 100 mm DTT and incubated for 15 min on ice. The plasma membranes were lysed with 10 μl of 10% Nonidet P-40 detergent. Supernatant was kept as cytoplasmic fraction (mESC CP). Insoluble nuclei were pelleted by centrifugation at 4,000 rpm for 5 min, washed briefly, and extracted by incubation with 1 ml of 1× nuclear extraction buffer (10× NEB = 20 mm Hepes, 1.5 mm MgCl2, 420 mm NaCl, 0.2 mm EDTA, pH 8.0, 25% glycerol (v/v)) with 2 μl of 100 mm DTT for 30 min at 5 °C with rotation. After centrifugation at 4,000 rpm, the supernatant was collected as nuclear extract (mESC NE); soluble protein was precipitated using ice-cold acetone overnight at −20 °C. The pellet was dried and solubilized in 40 μl of 8 m urea, 100 mm Tris, pH 7.8, at room temperature for ∼20 min. Reduction and alkylation were performed using 5 mm DTT for 30 min at 37 °C and 10 mm iodoacetamide for 30 min in the dark at 25 °C. 120 μl of 100 mm Tris, pH 7.8, and 1 mm CaCl2 (final concentration) was added before the addition of 2 μl of immobilized trypsin (Poroszyme; Applied Biosystems, Mississauga, Canada) for a 14-h incubation at 37 °C. Digestion was stopped by addition of formic acid to a 1% final concentration.
Yeast (strain S288C) soluble cell-free extract was generated by glass bead beating as described previously (23). 5 mg (500 μl of 10 mg/ml protein) of soluble extract was precipitated overnight at −20 °C by the addition of 5 volumes of ice-cold acetone. After washing twice with ice-cold acetone, the pellet was reconstituted in 200 μl of 50 mm ammonium bicarbonate, pH 7.8, and the final protein concentration was measured to be 5.94 μg/μl. 10 μg of trypsin (Roche; Mississauga, Canada) was added, and digestion was performed at 37 °C for 14 h. After addition of formic acid to 1%, the sample was stored at 5 °C prior to use.
LC-MS/MS Assay Development
The goal was to generate LC-MS/MS assays for both the ion trap and the triple quadrupole mass spectrometer. To this end, individual solubilized synthetic peptides were infused directly without further purification using either an EASY-nLC or a QuickQuan (Thermo Fisher) autosampler HPLC pump system to record optimal peptide fragmentation parameters on either an LTQ ion trap or a TSQ triple quadrupole tandem mass spectrometer (Thermo Fisher Scientific, San Jose, CA). MRM specific information was captured partially through direct infusion mass spectrometry and continuous isolation and fragmentation of the target precursor m/z at unit resolution. Full MS/MS scans were collected with predicted collision energy using Pinpoint (Thermo Fisher Scientific). To obtain optimal collision energies for each product ion, we used a QuickQuan autosampler which injected 200 μl of ∼100 ng/μl peptide at a flow rate of 100 μl/min. QuickQuan software then automatically selected the 10 most intense product ions to optimize collision energy ranging from 10 to 50 eV. In both cases, 10 top scans were extracted with in-house software and stored in a reference database. Selected subsets of the synthetic peptides were then mixed and spiked at various concentrations into digests of mESC nuclear extracts (5 μg) or yeast (1 μg) to record individual retention time relative to proteolytic background using nanoflow chromatography and, a column consisting of 5-cm × 250-μm inner diameter fused silica directly connected to a 15-cm × 75-μm inner diameter fused silica tubing packed with Luna C18 (3 μm) material (Phenomenex; Torrence, CA) terminating with a fine tip opening of ∼10 μm. A 45-min organic chromatography gradient (unless otherwise noted) was used to elute peptides off the column. Four “sentinel” internal retention time markers (spiked BSA peptides FKDLGEEHFK, HLVDEPQNLIK, KVPQVSTPTLVEVSR, and LVNELTEFAK) with limited sequence overlap to unrelated proteins were used to calculate relative retention time windows for each target peptide through the generation of a linear regression of recorded retention time values and actual retention time values as observed just prior to each analysis. An alignment using retention time marker peptides is shown in supplemental Fig. 2 and listed in supplemental Table 2.
LC-MS/MS Analysis of Dilution Series of Synthetic Peptides Spiked into Biological Samples
For the validation experiment, serial dilutions were generated of an unlabeled and heavy isotope (13C15N)-labeled synthetic peptide (FEALQLSLK) representing Pou5f1 (Oct4) at a 1:1 ratio (wt/wt) were spiked to a final concentration of 1 ng/μl, 100, 10, 1 pg/μl and 100 fg/μl into 1 μg of yeast digest. Then each serial dilution was analyzed in triplicate after loading onto a self-packed microcolumn (described above) using a Proxeon EASY-nLC autosampler and nanopump HPLC system (Proxeon, Odense, Denmark). Targeted LC-MS/MS in TPM mode (i.e. isolating and fragmenting target peptide precursors only (10) was performed using a chromatographic gradient (see supplemental Methods) with select target precursor ions (m/z 524.80, 528.80) continuously isolated and fragmented in a hybrid LTQ Orbitrap Velos instrument (Thermo Fisher Scientific) in addition to four sentinel precursor masses. Isolation width was kept at 2 m/z and normalized collision energy at 35, while allowing a maximum ion time of 100 ms to achieve ion trap targets of 1 × 105. A high resolution (R = 60,000) Orbitrap scan over the mass range of m/z 300–2000 was included in each cycle, followed by 6 collision-induced dissociation spectra in the Velos ion trap. Together with the Velos ion trap scans, typical cycle times of 1.2 s were achieved.
For the mESC sample analysis, serial dilution experiments (1,000, 100, 10, and 1 of fmol peptide targets) were performed for selected peptide mixtures both alone and after spiking into 5 μg of mESC soluble nuclear extract. In each case, an LTQ linear ion trap was operated with similar values as for the Velos (above) only that ion trap targets were kept at 3 × 104 but was also programmed to continuously fragment and monitor target peptide precursors to collect full MS/MS spectra (TPM mode).
For MRM, 1 pmol of peptides (maximal 6) from one target protein were first analyzed alone and then spiked into 5 μg of digested mESC cytoplasmic fraction applying an LC-MRM-MS, monitoring all b- and y-ions using 40-ms dwell time at unit resolution for Q1 and Q3 and a scan window of 1 m/z. Retention time was recorded for each target peptide. TCorr was applied to identify the target peptides (as described below). In addition, a “transition rank list” (supplemental Table 1) was created with the intensity of each product ion when spiked in target background. Also, a blank run was collected to capture background interferences.
In a second round, and using a new chromatographic column, only the two top ranking and unique transitions above the precursor m/z were selected for MRM in a multiplexed assay scheduled to monitor target transitions in a 4-min window around the expected retention time. Expected retention times were calculated using an initial run with sample and spiked retention time marker peptides (100 fmol of BSA in ∼5 μg of digested mESC CP) and alignment of those markers to previous runs carrying recorded retention time information of each peptide (here from LTQ-TPM runs). The retention time schedule of 87 peptides (174 transitions) is shown in supplemental Fig. 3. Then scheduled LC-MRM-MS for 5 μg of mESC CP and NE was performed using a 45-min organic gradient driven by the EASY-nLC system (as described above for LC-TPM-MS). Finally, injections were repeated with spiking all target peptides in a mixture of ∼5 μg of mESC NE or CP (where noted) as control. Also blank injections were performed with same assays to define background noise. All yeast- and mESC-related experiments were run starting from no (zero) spike-in control to the highest spike-in sample concentrations.
Spectral Processing, TCorr Library Searching and Quantification
For data processing, the RAW files were extracted into dat-files and then converted into dta-file format to perform TCorr library search (described below). Most abundant or most specific product ions generated by the respective TPM scan were filtered in Xcalibur v2.0.6 or Xcalibur v2.1 using m/z window of ±0.5 around the target ions for data from LTQ or LTQ-Velos Orbitrap, respectively. For quantification, extracted ion chromatograms were evaluated in Xcalibur v2.1. Gaussian smoothing over 7 points was applied and peak picking automatically applied using standard settings (i.e. baseline window = 40, area noise factor = 5 and peak noise factor = 10). When a peak was detected, the area under curve (AUC), peak height, and retention time of chromatographic apex were exported to an Excel spreadsheet for further processing. We previously showed (10) that quantification using peak height after applying peak smoothing allows quantification for equal linear dynamic range as when AUC is used. For precursor ion scan quantification, the same filter criteria were applied with the exception of a ±5 parts/million selection window to extract ion chromatograms (i.e. m/z 524.79–525.81 (light) and m/z 528.80–528.82 (heavy)) before exporting quantitative information to Excel for regression analysis to calculate standard curves.
A library of MS/MS spectra from 371 synthetic peptides of mouse stem cell regulatory proteins was established through selecting top scoring spectra using database search algorithm (such as SEQUEST). The 10 most consistent and best scoring spectra were stored to obtain a final number of 3,710 spectra. Experimental data interpretation was done using a specialized algorithm (12) that interprets intensity patterns of selected spectral features (i.e. b- and y-ions) of experimental spectra (within a 1-Da mass tolerance) compared with each of the annotated reference spectra in the library. A high correlation score (i.e. TCorr value of >0.95) was used for a given spectral pattern of preselected features to identify targets. The presence of 10 spectral copies in the library allowed the algorithm to perform more robustly for subtle differences in relative product ion abundance. A correlogram was created to plot all individual and consecutive spectral correlations along the chromatographic elution, wherein the x-axis indicates retention time and the y-axis reports the TCorr correlation value of an individual match. This visualization allowed high contrast of background noise (typically TCorr below 0.8) from known targets (TCorr >0.95).
For quantification we used either AUC or peak height, as for the benchmarking experiment in yeast. However, for multiplexed assays we preferred using peak height as indicated, due to potentially longer cycle times and fewer data points across chromatographic peak.
RESULTS
Building effective multiplexed LC-MS/MS assays requires meaningful information about the experimental behavior of the targets, including: (i) selection of unique peptide sequences that map unambiguously to a single protein (or isoform) of interest; (ii) knowledge of specific, intense “transitions” produced upon peptide fragmentation that are least likely to suffer interference from irrelevant biomolecules; (iii) tuned instrument settings (e.g. collision energy) to achieve optimal signal-to-noise; and (iv) well defined data acquisition criteria to enhance assay performance. Although suitable “proteotypic” peptides (24, 25), chromatographic properties (26, 27), and instrument settings can either be predicted (28, 29), deduced from proteomic datasets (30), or obtained from public proteomic databases (e.g. the GPM (31) or PeptideAtlas (32)), optimal experimental assay parameters must be determined empirically for individual instrument platforms and different samples.
Building Targeted LC-MS/MS Assays around Synthetic Peptide Arrays
The presented method, illustrated schematically in supplemental Fig. 4, addresses these requirements listed above in three main steps:
First, candidate peptides mapping uniquely to each of proteins of interest are synthesized on high density SPOT membranes using miniaturized solid phase Fmoc chemistry (see “Experimental Procedures”). In selected cases, stable isotope-labeled peptides are generated with heavy lysine (K*; 13C15N), or arginine (R*; 13C15N) C termini to validate and benchmark retention time windows and putative identifications using the TCorr library spectral matching measure of unlabeled peptides (light) as described below. The peptides are then analyzed extensively by MS/MS, both individually by direct infusion and after pooling individual peptides by LC separation, to generate a high quality spectral reference library with unique information about peptide fragmentation patterns and retention times. The arrays are applicable to both triple quadrupole and trap design mass spectrometers that have unique benefits, although screening is particularly straightforward using ion trap instruments.
Second, experimental LC-MS/MS procedures are designed to detect these same targets reliably in complex biological samples. Instrument methods are optimized based on the most intense and unambiguous product ions produced upon peptide fragmentation to produce standard curves with well defined limits of detection and quantitation. This includes TPM assays on ion traps (i.e. isolating and fragmenting target peptide precursors only) or highly paralleled MRM assays on triple quadrupole mass spectrometers (i.e. tracking the two most intense precursor-product ion transitions using empirically optimal collision energy settings).
Finally, the multiplexed screening assays are implemented on biological samples to detect and quantify endogenous proteins. We use a new peptide identification algorithm, termed TCorr (12), to identify target peptides from the recorded MS/MS spectra based on selected product ion intensity patterns corresponding to the corresponding reference library.
Proof-of-concept: Peptide Dilution Series Spiked into a Yeast Whole Cell Digest
A major aspect of our method is the generation and systematic experimental evaluation of a panel of synthetic reference peptides for assay development. We benchmarked the entire procedure by synthesizing mammalian peptide targets both in unlabeled (light) and stable isotope labeled (heavy) form. We spiked a 10-fold serial dilution of a 1:1 mixture of the heavy/light peptides into a yeast-soluble protein digest over a broad dynamic range of 1 pmol down to 100 amol (corresponding to ∼1 ng down to ∼100 fg of synthetic peptide) per 1 μg of yeast digest. The samples were then analyzed in triplicate by TPM (ion trap) and by high resolution precursor scanning (Orbitrap) concomitantly using an LTQ Orbitrap Velos hybrid tandem mass spectrometer followed by spectral interpretation with TCorr algorithm. A stringent TCorr cutoff score of >0.95 was used to identify the targets whereas abundance was defined by measuring the AUC and peak height of the monoisotopic (and most abundant) precursor ion peak extracted using a ±5 parts/million m/z window (see “Experimental Procedures”).
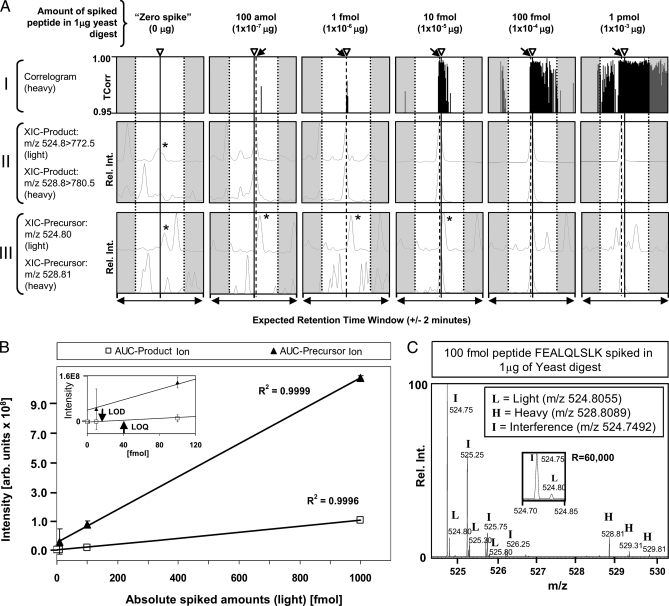
Plots (correlograms) of the TCorr values returned for the heavy isotope form of one representative target, FEALQLSLK, are shown in Fig. 1A, panel I. Whereas no significant TCorr signal was detected within a 4-min expected retention time window for the no spike-in control sample, matches were readily detected in the spike-in experiments (Fig. 1A, panels II and III). To define assay sensitivity formally, regression curve analysis was performed. Linearity (R2 > 0.99) was observed between 1 pmol and 1 fmol target spike-in levels, with a limit of detection (LOD; Fig. 1B) defined as significant deviation from “blank” (i.e.“zero-spike”) (38), of ∼13.1 fmol (∼13 pg) and limit of quantification (LOQ; Fig. 1B), defined as 3× LOD (38), of 39.3 fmol (∼40 pg), comparable with or even exceeding the corresponding precursor ion peak measurements (AUC) in high resolution full (MS1) scans (Fig. 1B and supplemental Figs. 5 and 6). Despite the presence of an abundant co-eluting and “co-fragmenting” isobaric interference (with accurate mass of m/z 524.7492) (Fig. 1C), the calculated coefficient of variation recorded for the target product ion intensity m/z 772.5 of the light target peptide (m/z 524.8055) abundance across three replicate measurements ranged from 0.014 up to 0.68 (or 1–68% without zero spike) over a broad dynamic range of 105, and from 0.010 to 0.46 (or 1 to 46%) for the heavy isotope reference, respectively (supplemental Table 3a). In addition, when calculating abundance ratios between the light/heavy forms, average coefficients of variation of 0.053 and 0.039 (or 5.3 and 3.9%) were observed for the light and heavy peptide forms (supplemental Table 3b). However, the actual fold change value was sometimes not predicted precisely and fold changes of about 2.0 and 0.5 were calculated instead of 1.0. When calculating the median across averaged spike-levels a 1.05-fold change was obtained for the light/heavy peptide abundance (supplemental Table 3b). Collectively, these results show that low abundance targets (1 fmol) can be confidently detected, and label-free relative quantification can be achieved based on quantification of product ion intensity once suitable assays have been built using synthetic reference peptides.
Fig. 1.
Proof-of-concept: LC-MS/MS analysis of a dilution series of light/heavy (1:1 ratio) synthetic Pou5f1 reference peptide FEALQLSLK spiked into digested yeast extract. A, panel I, correlograms showing TCorr values obtained for the isotope-labeled (heavy) target peptide at different spike levels at the expected target retention time window of ±2 min (estimated based on the sentinel marker peptides as described in supplemental methods). The arrowheads (and solid lines) indicate the center of the estimated retention time, whereas the arrows (and dashed lines) show the observed retention time (apex of chromatographic peak recorded for the heavy peptide) if different from estimated time. A, panel II, extracted ion chromatograms of the major product ions (m/z 772.50 and 780.50) generated by the light (unlabeled) and heavy target peptides (precursor m/z 524.8055 and 528.8089, respectively). A, panel III, extracted ion chromatograms of the target precursor ions (m/z 524.8055 and 528.8089), respectively, of high resolution spectra (R = 60,000) obtained in Orbitrap. The star (*) indicates an isobaric interference (precursor m/z 524.7492 or 524.75, rounded). B, calibration curve showing AUC values calculated for extracted ion chromatograms of the product ion m/z 772.5 (target peptide precursor m/z 524.8055) obtained on an LTQ-Velos ion trap (squares) and the corresponding precursor ion (m/z 524.8055) signal acquired in high resolution spectrum on an Orbitrap (filled triangles). Inset, zoom-in. Arrows mark the LOD of 13.1 fmol and LOQ of 39.3 fmol, respectively. C, zoom-in (m/z 524.30–530.30) of a high resolution full scan spectrum obtained on an Orbitrap of a 100 fmol spike-in experiment showing the isotope envelopes of both the light (L) and heavy (H) precursor ion target and a dominant isobaric (m/z 524.7492) interference (I). Inset, distinct monoisotopic peaks of the light target and the interference. All shown retention time windows were estimated using linear regression of four retention time marker peptides (here called sentinels), which were monitored throughout the experiment. The expected retention time is the linear extrapolation of retention times from the sentinel position in each LC/MS/MS run (for linear regression see supplemental Fig. 2 and supplemental Table 2).
Development and Application of Synthetic Arrays for Endogenous Protein Sequencing
To illustrate the process of using synthetic peptide arrays to monitor the endogenous components of biological pathways, below we describe the development and implementation of LC-MS/MS assays designed to investigate a regulatory network of 12 mouse proteins linked to mammalian stem cell renewal and pluripotency in nuclear extracts prepared from mESCs. Targets included components of the upstream JAK/STAT3 signaling cascade (21) (Table 1a) that interact directly or indirectly with transcription factors Pou5f1/Oct4, Sox2, c-Myc, and Klf4 (33) and related factors such as Nanog and Sall4 (34) (Table 1b) that modulate self-renewal and pluripotency.
Table I.
Selected proteins, accession numbers, protein class, and number of optimized peptides detected by LC-MRM-MS (a) and LC-TPM-MS (b)
| Protein | Swiss-Prot accession no. | Protein classa | Peptides predicted | Assay optimization | Successfully detected as endogenous |
|---|---|---|---|---|---|
| a. LC-MRM | |||||
| Jak1 | P52332 | K | 22 | 22 | 3 |
| Stat1 | P42225 | TF | 6 | 6 | 1 |
| Stat3 | P42227 | TF | 13 | 13 | 1 |
| Lifr | P42703 | R | 3 | 3 | 3 |
| Akt1 | P31750 | K | 15 | 13 | 5 |
| P85A | P26450 | K | 31 | 30 | 3 |
| Total peptides | 90 | 87 | 16 | ||
| b. LC-TPM | |||||
| Pou5f1 | P20263 | TF | 6 | 6 | 1 |
| Sox2 | P48432 | TF | 6 | 5 | 1 |
| Klf4 | Q60793 | TF | 1 | 1 | 0 |
| c-Myc | P01108 | TF | 3 | 3 | 0 |
| Nanog | Q80Z64 | TF | 1 | 1 | 0 |
| Sall4 | Q8BX22 | TF | 7 | 6 | 1 |
| Total peptides | 24 | 22 | 3 |
TF, transcription factor; R, receptor; K, kinase.
Step 1: Target Selection, Microscale Synthesis, and Feature Extraction
An initial challenge was to define suitable peptides for synthesis given that only a subset of the targets chosen has been previously detected in stem cells by LC-MS/MS. For example, public repositories, e.g. GPM (www.thegpm.org), PeptideAtlas (www.peptideatlas.org), and Peptidome (www.ncbi.nlm.nih.gov/projects/peptidedome), curated only a single peptide for both Sox2 and Pou5f1 (supplemental Fig. 7A). To increase the odds of detection, we opted to synthesize multiple unique (5–10 peptides, where possible) tryptic sequences per protein (supplemental Table 1) using generic selection criteria (see “Experimental Procedures” and supplemental Methods for details). A total of 384 distinct peptides (some in duplicate) were programmed for synthesis on a single SPOT membrane resulting in 371 unique peptide sequences.
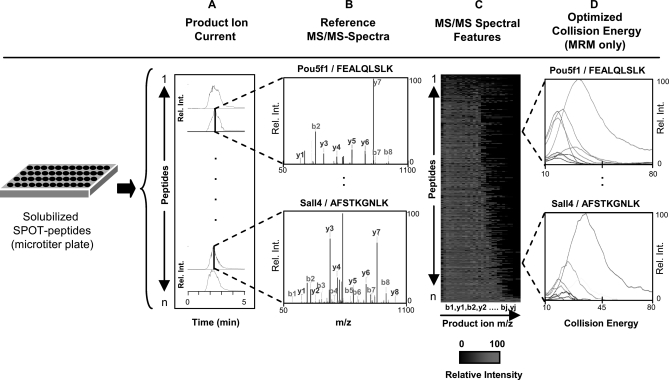
Target synthesis and purity were first verified in batch mode by direct infusion nanoelectrospray MS/MS analysis using a linear ion trap tandem mass spectrometer (see supplemental Methods). The resulting product ion patterns (Fig. 2, A and B) were validated by using SEQUEST algorithm against a database corresponding to the intended target sequences (371 peptides) and, as a more stringent test, against all annotated mouse proteins (see “Experimental Procedures”). More than 90% of the peptides were positively identified, most with a predominant +2 charge state. Ten representative highest scoring (by XCorr) doubly charged MS/MS spectra were then extracted to create a reference spectral library (Fig. 2, B and C). In total, only 4 of the 371 candidates attempted did not result in an interpretable fragmentation pattern, whereas another 24 produced low ion current and so were flagged not to be selected for further assays. Hence, despite the simplicity of the peptide selection rules, the SPOT peptides were almost uniformly of good quality with estimated 80–90% purity. Moreover, estimates by LC/MS (JPT Peptide Technologies) and quantification using external standard curve of the high purity SLG tag by us (see supplemental Fig. 1B and supplemental Methods) suggest an average yield of ∼50 nmol (∼5–10 μg), far exceeding the amount needed to optimize experimental conditions for LC-MS/MS assay development.
Fig. 2.
Generation of a high quality reference MS/MS-spectral library. A, solubilized synthetic peptides are analyzed by electrospray ionization, with targeted isolation and fragmentation using an ion trap or triple quadrupole mass spectrometer and the product ions recorded continuously over 5 min. B, collected MS/MS spectra sequence-verified by database searching using SEQUEST. C, characteristic experimental intensity patterns of distinguishing b- and y-ion features stored in a relational database. D, MRM assay development using empirically optimized collision energy settings, shown here based on the breakdown curves obtained for the 10 most intense transitions (see “Experimental Procedures” and supplemental Methods” for details).
As MRM is an increasingly popular platform for multiplexed assays monitoring target peptides, we defined triple quadrupole instrument parameters that produce high responding product ions suitable for target quantification by recording the signal intensities of all predicted b- and y-ions while incrementally ramping collision energy but not tube lens voltages (see supplemental Methods and supplemental Table 1). A heat map of the summed product ion intensity patterns produced by each of the synthetic peptides is shown Fig. 2C, whereas Fig. 2D shows the results of individual transitions obtained for two representative targets (FEALQLSLK, from Pou5f1 as before; and AFSTKGNLK, from Sall4). The optimal instrument settings (i.e. collision energies) and corresponding target intensity profiles were also stored in the spectral database.
Step 2: Constructing Multiplexed LC-MS/MS Assays
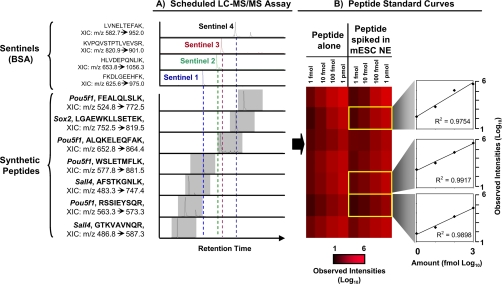
Analytical nanoflow LC is prone to column-to-column retention time fluctuations that can affect long term assay robustness (35). We therefore devised a scheduling strategy exploiting the characteristic recorded chromatographic retention times of the synthetic peptides relative to those of a common set of chromatographic markers, termed sentinels, that can be sensitively and reproducibly detected when spiked into diverse biological samples to build robust nanoflow assays with high target density and detection efficacy (see “Experimental Procedures”). Narrow predefined data acquisition windows were then defined based on normalization of target retention times relative to the sentinels (gray highlights in Fig. 3A). Once relative retention time to sentinels was recorded, alignment was performed using linear regression analysis of stored sentinel retention times to “actual retention time” in current LC-MS/MS runs to find expected retention times for each target peptide (supplemental Fig. 2 and supplemental Table 2).
Fig. 3.
Multiplex assay development. A, representative scheduled LC-MS/MS assay monitoring multiple reporter peptides corresponding to transcription factors regulating embryonic stem cell fate. Four exogenous sentinel marker peptides are jointly monitored in parallel to calculate relative retention time (stored in relational database) to control target data acquisition windows (gray highlights). The XIC of the most intense product ions of each peptide is indicated. B, heat map showing ion abundances for peptide standard curves generated using synthetic peptides alone or after spiking of the reference standards into a digested mESC NE. The zoom-ins show selectively linear signal response over 3 orders of magnitude (10-fold dilutions from 1 pmol to 1 fmol) using log-log (base 10) plots.
Standard 10-fold serial dilution experiments were then performed with the synthetic peptides alone and after spiking into the mESC nuclear extract (see “Experimental Procedures”). Fig. 3B shows measured chromatographic peak heights of the most intense product ions of seven representative peptide targets of three pluripotency markers. Although several peptides were only detectable at the highest concentration, most showed good linearity (average R2 of >0.94 in log-log plots) across a range of 3 orders of magnitude, as exemplified by peptides LGAEWKLLSETEK (Sox2), AFSTKGNLK (Sall4), and RSSIEYSQR (Pou5f1) (Fig. 3B and supplemental Fig. 8). Nevertheless, since high abundance components of biological matrices can potentially confound assay accuracy (36), spectral features have to be evaluated rigorously to ensure specificity when detecting low abundance targets (37–40). Although often only a few (3–8) transitions are commonly used in the literature to track multiple targets to achieve sufficient assay sampling efficiency (1, 39, 41), it is currently unclear how many product ions are minimally needed to define endogenous target identity unambiguously (38, 41). Hence, we first opted to collect full MS/MS scans using a fast scanning linear ion trap followed by pseudo-MRM-like extraction of b- and y-product ions that uniquely identify a target using a dedicated scoring function, TCorr (“Transition-Correlation”) (12). TCorr calculates the dot-product (or optionally the Pearson correlation) between the experimental b- and y-ion signal intensity patterns compared with the spectra reference library (Step 1). Precursor-product ion transitions with matching correlation values of 0.95 or greater to an annotated reference at the expected chromatographic retention time are deemed significant (12).
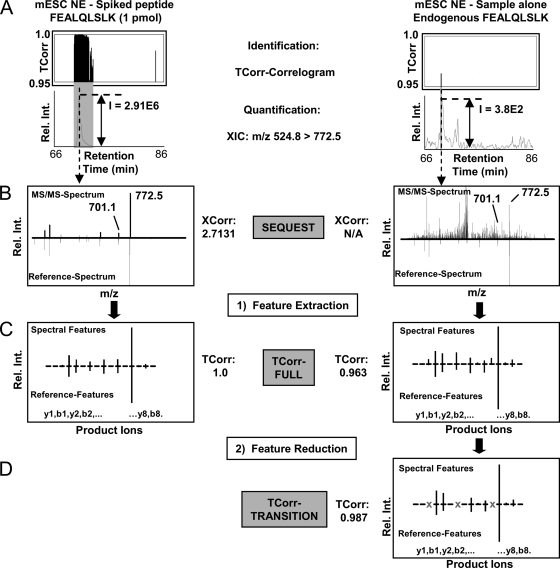
Fig. 4A (top) shows the TCorr values obtained across the chromatographic gradient (i.e.“correlogram”) for a representative Pou5f1 peptide, FEALQLSLK, for 1 pmol spike-in level and for the endogenous factor. Correlation values near 1.0 precisely define a single relevant peak at the expected target retention time window. The extracted ion current (XIC; Fig. 4A, bottom) of the most intense product ion (m/z 772.5) consistently gave the highest signal response (i.e. target could be measured at the lowest concentrations within the sample matrix) compared with either the precursor (m/z 524.8) or other product ions (supplemental Fig. 9). However, the conditional coexistence of other product ions with varied intensity generated the most significant correlation values such that TCorr outperformed detection of low abundance targets in terms of specificity and signal/noise compared with the XIC of the precursor or the most intense product ion alone (Fig. 4, B and C). Moreover, despite spurious peaks, the identity of the endogenous target could be unambiguously confirmed using TCorr based on the excellent match of the relevant product ion features to the curated spectra reference library (TCorr score = 0.963; Fig. 4C). Moreover, removal of potentially interfering product ion features improved TCorr score (TCorr score = 0.987; Fig. 4D).
Fig. 4.
Peptide identification and quantification using TCorr. A, representative correlograms showing the chromatographic profile of TCorr scores (0. 95 and higher) calculated for experimental MS/MS spectra recorded for spiked synthetic (left) or endogenous (right) Pou5f1 peptide FEALQLSLK in mESC NE matched against the corresponding reference spectral library over a specified retention time window 66- 86 min. Significant similarity in the observed b- and y-product ion intensity patterns occurs regardless of background interferences, allowing the XIC of the most intense target precursor-to-product ion transition to be reliably quantified. The gray highlights correspond to high confidence matches represented by the highest consecutive TCorr correlation scores. B, comparison of exemplar MS/MS spectra obtained for spiked and endogenous peptide (top) versus the corresponding library reference (bottom). XCorr correlation scores of a SEQUEST database search are indicated and obtained for the correct sequence FEALQLSLK. C, representative relative peak intensities of all b/y-ion transitions (TCorr-FULL) and TCorr-correlation score of 0.963 are shown. D, improved TCorr score 0.987 for endogenous peptide based on selected transitions freed from potential interferences (TCorr-TRANSITION).
Likewise, as for the LC-TMP-MS, we constructed LC-MRM-MS assays based on synthetic peptides. First, individual peptides were injected by direct infusion into the TSQ Access mass spectrometer by EASY-nLC system (used without the LC-column) and QuickQuan platform to record all b- and y-product ion intensities and the 10 most abundant ions, respectively. For the latter, optimization of parameters is automated, such as finding optimal collision energies (and tube lens parameters, here disabled) and product ions are already ranked upon acquisition and stored in a reference library (Excel file). For almost all cases, the top 10 ion intensities matched the ranking obtained from MS/MS spectra in the LTQ. However, as the next step we used EASY-nLC coupled to a microcolumn (same as for LC-TPM-MS) to inject mixtures of six synthetic peptides (∼5 pmol each) while monitoring all b- and y-ion transitions during a 45-min gradient and used predicted collision energies for entire peptides using Pinpoint 1.0. Peptide mixtures were run alone and spiked into ∼5-μg mESC cytoplasmic fraction. About 15 LC-MRM-MS runs monitoring all b- and y-ions were performed twice for a total of 87 synthetic peptides.
Step 3: Assay Implementation (Target Detection and Quantification)
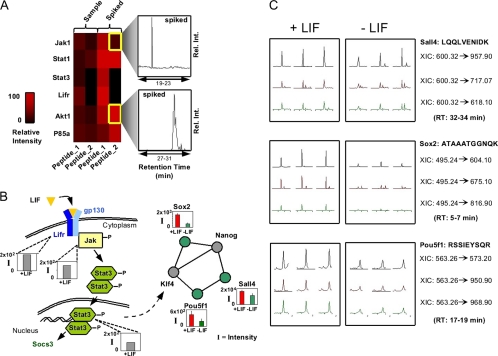
We used multiplexed MRM for top two ranking transitions of at least two peptides/protein (where possible) to examine the components of the JAK/STAT pathway activated by the LIF ligand (87 target peptides mapping to the transcription factors STAT1 and STAT3, the LIF receptor LIFR, and the protein kinases JAK1, Akt1, and P85a) in a nuclear extract prepared from ∼500,000 mESCs treated with LIF to maintain pluripotency. Sentinel-based scheduling, with and without spiking of the synthetic references, allowed target time window of minimally 4 min over a 45-min chromatographic gradient. Background noise was assessed and subtracted by monitoring an off-target signal recorded 5 min prior to the predicted target retention times. Fig. 5A shows an abundance heat map for representative target peptides (in Table 1a). The typical sampling routine for these assays was implemented such to inject (i) water (“blank,” to find background), (ii) mESC nuclear extract or cytoplasmic fraction (“sample”), and (iii) mESC cytoplasmic fraction spiked with >100 fmol of synthetic peptides each (“spike”). For data interpretation peak height was used at expected retention times within a tolerance of ±1 min.
Fig. 5.
Multiplexed quantitative assays of pluripotency pathway components. A, heat map showing estimated protein abundance of Jak1, STAT3, STAT1, Akt1, and LIFR in cytoplasmic extracts from pluripotent mESC populations based on two endogenous or spiked peptides (1 and 2) measured by MRM. Inset, representative (untransformed) relative intensity (y axis). B, pathway schematic showing experimental XIC values (I, intensity) obtained for peptides corresponding to endogenous Sall4, Pou5f1, and Sox2 levels in nuclear and cytoplasmic extracts from +LIF (self-renewal; red) and −LIF (differentiation; green) treated mESCs. C, actual protein abundance measurements of pluripotency factors in +LIF and −LIF mESC NEs as obtained by targeted LC-MS/MS experiments (TPM) on an LTQ-ion trap.
Next, to increase specificity through monitoring all product ions, we applied multiplex TPM mode assays to monitor changes in the levels of six core pluripotency factors (Pou5f1/Oct4, Sox2, cMyc, Nanog, Klf4, and Sall4; Table 1b) in nuclear extracts prepared from mESC grown with LIF (self-renewing) or after growth factor removal to initiate differentiation (see supplemental Methods). Again, soluble protein from about 500,000 cells was digested and quantified by three repeat measurements of the peak height of chromatographic peaks corresponding to the XIC of the most intense product ion above the precursor m/z across all experiments (Fig. 5B). Protein abundance decreases significantly (p value <0.01 by two-tailed t test) in Sall4, Sox2, and Pou5f1 upon differentiation (Fig. 5C; see supplemental Table 4 for details, with confirmation spectra shown in supplemental Figs. 10 and 11). Although these results are consistent with biological expectation, this analysis represents the first joint quantification of key nodes in a conserved regulatory pathway simultaneously during mESC fate changes by targeted LC-MS/MS. Although we anticipated detection of all proteins listed in Table 1b, Klf4 and Nanog were represented in our spectral library by only one tryptic peptide and hence might have been remained undetected in both samples. For example Klf4 and c-Myc were detected by data-dependent sampling using extensive prefractionation techniques by different peptides than selected here (data not shown).
DISCUSSION
We have presented a targeted proteomics approach that enables the routine development and implementation of sensitive LC-MS/MS assays to monitor and quantify multiple components of critical biological pathways in a single experiment. Our approach is based on the systematic generation and empirical evaluation of synthetic peptide arrays to generate high confidence reference information to fine tune experimental parameters providing the most informative and intense fragment ions by which to detect specific protein targets. Our method builds on targeted LC-MS/MS procedures (i.e. TPM/MRM) that allow for more sensitive, consistent, and quantitative detection of low abundance proteins than global proteome profiling (10, 37). Although the use of synthetic peptides is not new (42, 43), we have established that microscale SPOT membrane synthesis offers a particularly rapid and cost-effective means of creating many peptide standards, both in native and heavy isotope-labeled form, good enough to generate high quality MS/MS reference spectra and to use for multiplex assay design and as internal standards for data interpretation (47). Through automation of each step, multiplexed assays can be established in 1–2 weeks at moderate cost for virtually any pathway, including proteins never detected previously by MS/MS. Using this approach, we showed that the expression of known pluripotency factors in mESC show down-regulation in a time frame of at least 48 h withholding LIF, in congruency with previous studies. The ability to track pathway component levels in a single assay should pave the way for novel insight into the mechanisms of emergent cell behaviors such as reprogramming and self-renewal. Similar assays could be used to investigate the dynamics of cellular signaling cascades, to confirm gene silencing knock-down (i.e. RNAi) experiments at the protein level, or to evaluate multiple candidate disease biomarkers comparatively for personalized medicine.
Suitable reporter peptides for assay development can be selected either by prior knowledge (e.g. using proteotypic peptides documented in public databases such as GPM or PeptideAtlas) or by prediction. The former represent species that are proven to be detectable but are restricted to previously characterized proteins and certain biological contexts. Since many signaling factors have yet to be documented experimentally, computational algorithms (24, 25, 58) offer a promising alternate approach to select high responding candidates for targeted proteomic screens of pathways. Although we opted to apply simple generic filters to select reporter peptides, our systematic empirical optimization strategy allows for the evaluation of peptide ionizability, chromatographic separability, and identifyability in complex biological mixtures to narrow a final set of suitable assay candidates. Although models of peptide retention time have been reported (26, 27), we demonstrated that the use of a chromatographic sentinel markers greatly facilitates the building of efficient multiplexed assays that can be transferred between platforms or interday analyses (with a priori prediction). Although chromatographic peaks occasionally fluctuate by up to 2 min, we commonly achieved a prediction precision of ±6 s (±0.11 min with post hoc prediction) across more than 20 runs using 2 different columns over multiple days of analysis (supplemental Table 2). We also tested the accuracy of retention time prediction based on the predicted hydrophobicity of peptides and compared these with the observed retention times of ∼118 synthetic peptides used in our experiments through linear regression analysis. Although an overall correlation was observed (supplemental Fig. 12), individual peptide retention times often deviated considerably (i.e. several minutes) from the predicted values. However, one drawback is that adjustment of larger retention time fluctuations (>3 min) has to be done manually by correcting predicted retention time windows. Hence, assay design would gain further robustness if the prediction of estimated retention time windows could be done in real time during the LC-MS/MS run, by triggering target analysis based on detection of a sentinel or another high abundant marker peptide in a precursor scan, similar to a method reported by Jaffe et al. (44).
A key, but often underappreciated, aspect for targeted proteomic analysis is the selection of spectral features to confirm target identity rigorously. In the past, the criteria for target identification have ranged from the co-elution of three to eight distinct transitions (41) up to the collection of full mass range MS/MS spectra triggered by a single transition (45). Since the MS/MS spectra of low abundant peptides are often confounded by product ions from irrelevant higher abundant isobaric peptides (46), we calculated the correlation between preselected b- and y-ions using relative product ion intensity patterns to minimize the effects of spurious interferences. We found that TCorr-based feature comparisons of spectra acquired at the appropriate target retention windows can result in more high confidence detection of low abundance protein in stem cell extracts compared with more conventional identification criteria (47, 48). Abatiello et al. recently presented the AuDIT algorithm (49) to critically select most concise MRM transitions as a measure for reliable detection and identification, and Prakash et al. recently showed that scoring based on extracted transition ion intensities can enhance performance (13), but we are unaware of other library search algorithms which implement chromatographically aligned intensity pattern correlation to confirm target identity based on few product ion features in a complex sample and not based on whole MS/MS spectrum interpretation.
With the increasing usage of target-driven (i.e. MRM-type) experiments, target identification in extremely complex sample matrices, such as mammalian cell extracts, based on only a few spectral features is prone to errors, especially when an even smaller subset of features is used to rank known product ion intensities. To this end, we have previously examined the influence of database size on the identification of target peptides based on selected product ion intensities by Monte-Carlo simulation (12). The results showed that when a set of more than eight transitions is used, the influence of database size on matching performance is mitigated. High resolution MS/MS spectra might help in selected cases to remove product ion interference or increase the specificity of pattern matching algorithms. We found that peptide fragments generated in an ion trap and Orbitrap higher energy collisional dissociation cell are usually very similar (supplemental Fig. 13). Nevertheless, monitoring only a few transitions is already beneficial for minimizing interferences, making MRM-type experiments (on both triple quadrupoles and traps) powerful tools for detecting low abundance species. Accordingly, we observed a slightly improved TCorr score (from 0.963 to 0.987) when removing three potential interfering product ions from consideration (Fig. 4D). Similarly, in our proof-of-concept benchmarking experiment using heavy labeled internal standards spiked into a yeast digest as a complex proteolytic background, we observed ∼10× lower LOD with filtering one selected subset of interfering product ions (supplemental Figs. 5 and 6).
Furthermore, calibrated assays using serial dilutions of spiked peptides into proteolytic background can be used to calculate the amount of molecules per cell. In our case for the analysis of target peptides in ∼500,000 cells (Fig. 5B) we can calculate the detection of at least ∼1,248 molecules/cell using the proximal amount of 1 fmol (∼1 pg) peptide spiked (e.g. AFSTKGNLK, 965 g/mol) in the cell lysate (see supplemental Methods for calculation).
To move from global proteomic discovery profiling to hypothesis-driven quantitative assessments, high density assays must be built using optimal empirical parameters and instrument settings with adequate sampling rates to ensure accurate target quantification. Although MRM on triple quadrupole instruments is more quantitative (10) in this respect, implementation of TPM (PIM) style LC-MS/MS screening on trap-type instruments is potentially more straightforward as only two experimental parameters need to be generated (i.e. reference MS/MS spectra and knowledge of peptide retention time). For example, Sullivan et al. (50) showed previously that target glycopeptides could be detected at low abundance levels using ion traps. Another benefit of TPM is that it can generate MS/MS spectra quickly (typically ∼100 ms) with comparable sensitivity as for only few selected transitions by MRM. Table 1a showed data acquired by MRM, and Table 1b for TPM for different datasets confirmed the suitability of both platforms. Moreover, product ion spectra for extracted b- and y-ions have similar abundance patterns (supplemental Fig. 7, B and C) with the tendency of ion trap spectra being more reproducible in their relative product ion intensities. However, a detailed comparison of instrument performance for targeted analysis showing comparable sensitivity for both the ion trap and triple quadrupole mass spectrometers can be found in previous publication by Sandhu et al. (10).
Although stable isotope-labeled reference peptide arrays could be exploited to increase the accuracy of protein quantification (e.g. AQUA (43)), we show that comparable results can be achieved by careful implementation of a “label-free” quantification strategy based on measuring the intensities of dominant product ions normalized to individual peptide standard curves. Indeed, we have shown that our TPM assays provide a linear dynamic range over at least 3 orders of magnitude (1 ng down to 1 pg target/μg of yeast digest; Fig. 1B) for carefully chosen product ions. The relatively small coefficient of variation reported here for the proof-of-concept experiment (supplemental Table 3b) gives reason to use only one most unique (and most abundant) product ion to quantify a peptide. However, statistical error can be calculated by using replicate injections instead to find statistical evidence for a differential expression between samples. We note that the use of heavy isotope-labeled peptides is also helpful in finding the pairing unlabeled peptide species and confirming relative quantification levels (43, 51, 52). Software for aligning and integrating multiple LC-MS/MS datasets (53–56) could be used to increase quantification accuracy and detection sensitivity further, whereas modest sample prefractionation can be used to enhance detection limits (57).
We would like to point out that with our demonstrated method it should be possible to obtain sequence information of multiple peptides of interest in real time, during sample acquisition. TCorr could be tied directly into mass spectrometer instrument software to obtain sequence interpretation on the fly. However, the limitation is that synthetic peptides have to be sequenced a priori to generate an “absolute” reference spectrum. Nevertheless, given the fact that high throughput peptide synthesis can build high quality reference libraries quickly, this limitation might not be as significant any more. An important consideration is sampling speed when larger peptide sets are monitored. The spectral density within short cycle times is crucial for capturing reliable quantitative information for proteome scale experiments with, e.g. ultra-HPLC. Hence, faster mass spectrometers using novel geometries are needed for ultrafast sampling by maintaining good ion statistics. Nevertheless, these developments will benefit the analyses of large numbers of clinical samples with limited availability where one wants to access large scale quantitative proteome sequence information in real time.
Supplementary Material
Acknowledgments
We thank members of the Emili and Zandstra laboratories for advice, assistance, and critical suggestions. We thank Thomas Kislinger and Andrei Starostin for the contribution of Orbitrap data, Pierre Havugimana for analyzing the mESC nuclear extracts using prefractionation, Carl White for feedback on the manuscript, and Mike MacCoss for providing the P3-predictor software (predecessor of the Pinpoint offered by Thermo Fisher) to predict collision energies and precalculate suitable MRM transitions.
Footnotes
* This work was supported by funds from the Canadian Foundation for Innovation, the Ontario Research Fund/Ontario Ministry for Innovation, and an National Sciences and Engineering Research Council grant and Canadian Institutes of Health Research Grant MOP-57885 (to P. W. Z.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1–4, Figs. 1–13, and Methods.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables 1–4, Figs. 1–13, and Methods.
1 The abbreviations used are:
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- AUC
- area under curve
- CP
- cytoplasmic fraction
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- LIF
- leukemia inhibitory factor
- LOD
- limit of detection
- LOQ
- limit of quantification
- mESC
- mouse embryonic stem cell
- MRM
- multiple reaction monitoring
- NE
- nuclear extract
- TPM
- targeted peptide monitoring
- XIC
- extracted ion chromatogram
- SVM
- support vector machine.
REFERENCES
- 1.Anderson L., Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 2.Rifai N., Gillette M. A., Carr S. A. (2006) Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 [DOI] [PubMed] [Google Scholar]
- 3.Ahrens C. H., Brunner E., Hafen E., Aebersold R., Basler K. (2007) A proteome catalog of Drosophila melanogaster: An essential resource for targeted quantitative proteomics. Fly 1, 182–186 [DOI] [PubMed] [Google Scholar]
- 4.Kislinger T., Gramolini A. O., MacLennan D. H., Emili A. (2005) Multidimensional protein identification technology (MudPIT): Technical overview of a profiling method optimized for the comprehensive proteomic investigation of normal and diseased heart tissue. J. Am. Soc. Mass Spectrom. 16, 1207–1220 [DOI] [PubMed] [Google Scholar]
- 5.Eng J. K., McCormack A., Yates J. R. I. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 6.Marcotte E. M. (2007) How do shotgun proteomics algorithms identify proteins? Nat. Biotechnol. 25, 755–757 [DOI] [PubMed] [Google Scholar]
- 7.Isserlin R., Emili A. (2008) Interpretation of large-scale quantitative shotgun proteomic profiles for biomarker discovery. Curr. Opin. Mol. Ther. 10, 231–242 [PubMed] [Google Scholar]
- 8.Kulasingam V., Smith C. R., Batruch I., Buckler A., Jeffery D. A., Diamandis E. P. (2008) “Product ion monitoring” assay for prostate-specific antigen in serum using a linear ion-trap. J. Proteome Res. 7, 640–647 [DOI] [PubMed] [Google Scholar]
- 9.Lange V., Malmström J. A., Didion J., King N. L., Johansson B. P., Schäfer J., Rameseder J., Wong C. H., Deutsch E. W., Brusniak M. Y., Bühlmann P., Björck L., Domon B., Aebersold R. (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol. Cell. Proteomics 7, 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandhu C., Hewel J. A., Badis G., Talukder S., Liu J., Hughes T. R., Emili A. (2008) Evaluation of data-dependent versus targeted shotgun proteomic approaches for monitoring transcription factor expression in breast cancer. J. Proteome Res. 7, 1529–1541 [DOI] [PubMed] [Google Scholar]
- 11.Schmidt A., Gehlenborg N., Bodenmiller B., Mueller L. N., Campbell D., Mueller M., Aebersold R., Domon B. (2008) An integrated, directed mass spectrometric approach for in-depth characterization of complex peptide mixtures. Mol. Cell. Proteomics 7, 2138–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J., Hewel J. A., Fong V., Chan-Shen-Yue M., Emili A. (2009) Critical evaluation of product ion selection and spectral correlation analysis for biomarker screening using targeted peptide multiple reaction monitoring. Clin. Proteomics 5, 3–14 [Google Scholar]
- 13.Prakash A., Tomazela D. M., Frewen B., Maclean B., Merrihew G., Peterman S., Maccoss M. J. (2009) Expediting the development of targeted SRM assays: Using data from shotgun proteomics to automate method development. J. Proteome Res. 8, 2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuster B., Schirle M., Mallick P., Aebersold R. (2005) Scoring proteomes with proteotypic peptide probes. Nat. Rev. Mol. Cell Biol. 6, 577–583 [DOI] [PubMed] [Google Scholar]
- 15.Anderson N. L., Anderson N. G., Pearson T. W., Borchers C. H., Paulovich A. G., Patterson S. D., Gillette M., Aebersold R., Carr S. A. (2009) A human proteome detection and quantitation project: hPDQ. Mol. Cell. Proteomics 8, 883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank R. (2002) The SPOT synthesis technique. Synthetic peptide arrays on membrane supports–principles and applications. J. Immunol. Methods 267, 13–26 [DOI] [PubMed] [Google Scholar]
- 17.Frank R., Overwin H. (1996) SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol. Biol. 66, 149–169 [DOI] [PubMed] [Google Scholar]
- 18.Gausepohl H., Boulin C., Kraft M., Frank R. W. (1992) Automated multiple peptide synthesis. Pept. Res. 5, 315–320 [PubMed] [Google Scholar]
- 19.Hilpert K., Winkler D. F., Hancock R. E. (2007) Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2, 1333–1349 [DOI] [PubMed] [Google Scholar]
- 20.Volkmer R. (2009) Synthesis and application of peptide arrays: Quo vadis SPOT technology. Chembiochem 10, 1431–1442 [DOI] [PubMed] [Google Scholar]
- 21.Davey R. E., Onishi K., Mahdavi A., Zandstra P. W. (2007) LIF-mediated control of embryonic stem cell self-renewal emerges due to an autoregulatory loop. FASEB J. 21, 2020–2032 [DOI] [PubMed] [Google Scholar]
- 22.Wenschuh H., Volkmer-Engert R., Schmidt M., Schulz M., Schneider-Mergener J., Reineke U. (2000) Coherent membrane supports for parallel microsynthesis and screening of bioactive peptides. Biopolymers 55, 188–206 [DOI] [PubMed] [Google Scholar]
- 23.Lam M. H., Urban-Grimal D., Bugnicourt A., Greenblatt J. F., Haguenauer-Tsapis R., Emili A. (2009) Interaction of the deubiquitinating enzyme Ubp2 and the E3 ligase Rsp5 is required for transporter/receptor sorting in the multivesicular body pathway. PLoS One 4, e4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fusaro V. A., Mani D. R., Mesirov J. P., Carr S. A. (2009) Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat. Biotechnol. 27, 190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallick P., Schirle M., Chen S. S., Flory M. R., Lee H., Martin D., Ranish J., Raught B., Schmitt R., Werner T., Kuster B., Aebersold R. (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 [DOI] [PubMed] [Google Scholar]
- 26.Baczek T., Kaliszan R. (2009) Predictions of peptides' retention times in reversed-phase liquid chromatography as a new supportive tool to improve protein identification in proteomics. Proteomics 9, 835–847 [DOI] [PubMed] [Google Scholar]
- 27.Klammer A. A., Yi X., MacCoss M. J., Noble W. S. (2007) Improving tandem mass spectrum identification using peptide retention time prediction across diverse chromatography conditions. Anal. Chem. 79, 6111–6118 [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z. (2004) Prediction of low-energy collision-induced dissociation spectra of peptides. Anal. Chem. 76, 3908–3922 [DOI] [PubMed] [Google Scholar]
- 29.Klammer A. A., Reynolds S. M., Bilmes J. A., MacCoss M. J., Noble W. S. (2008) Modeling peptide fragmentation with dynamic Bayesian networks for peptide identification. Bioinformatics 24, i348–i356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kline K. G., Frewen B., Bristow M. R., Maccoss M. J., Wu C. C. (2008) High quality catalog of proteotypic peptides from human heart. J. Proteome Res. 7, 5055–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craig R., Cortens J. C., Fenyo D., Beavis R. C. (2006) Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 5, 1843–1849 [DOI] [PubMed] [Google Scholar]
- 32.Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: A resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 34.Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 35.Kiebel G. R., Auberry K. J., Jaitly N., Clark D. A., Monroe M. E., Peterson E. S., Toliæ N., Anderson G. A., Smith R. D. (2006) PRISM: A data management system for high-throughput proteomics. Proteomics 6, 1783–1790 [DOI] [PubMed] [Google Scholar]
- 36.Picotti P., Aebersold R., Domon B. (2007) The implications of proteolytic background for shotgun proteomics. Mol. Cell. Proteomics 6, 1589–1598 [DOI] [PubMed] [Google Scholar]
- 37.Picotti P., Lam H., Campbell D., Deutsch E. W., Mirzaei H., Ranish J., Domon B., Aebersold R. (2008) A database of mass spectrometric assays for the yeast proteome. Nat. Methods 5, 913–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keshishian H., Addona T., Burgess M., Kuhn E., Carr S. A. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keshishian H., Addona T., Burgess M., Mani D. R., Shi X., Kuhn E., Sabatine M. S., Gerszten R. E., Carr S. A. (2009) Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 8, 2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson N. L., Anderson N. G., Haines L. R., Hardie D. B., Olafson R. W., Pearson T. W. (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res. 3, 235–244 [DOI] [PubMed] [Google Scholar]
- 43.Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaffe J. D., Keshishian H., Chang B., Addona T. A., Gillette M. A., Carr S. A. (2008) Accurate inclusion mass screening: A bridge from unbiased discovery to targeted assay development for biomarker verification. Mol. Cell. Proteomics 7, 1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lenz C., Kühn-Hölsken E., Urlaub H. (2007) Detection of protein-RNA crosslinks by NanoLC-ESI-MS/MS using precursor ion scanning and multiple reaction monitoring (MRM) experiments. J. Am. Soc. Mass Spectrom. 18, 869–881 [DOI] [PubMed] [Google Scholar]
- 46.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 47.Hewel J., Liu J., Emili A. (2008) High-resolution biomarker discovery: From global proteomic profiling to targeted tandem mass spectrometry assays, in 56th American Society of Mass Spectrometry Conference, Denver CO, June 1–5 [Google Scholar]
- 48.Hewel J., Liu J., Emili A. (2008) Targeted proteomics of transcription factors. Seventh Human Proteome Organisation Conference, Amsterdam, The Netherlands [Google Scholar]
- 49.Abbatiello S. E., Mani D. R., Keshishian H., Carr S. A. (2010) Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin. Chem. 56, 291–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan B., Addona T. A., Carr S. A. (2004) Selective detection of glycopeptides on ion trap mass spectrometers. Anal. Chem. 76, 3112–3118 [DOI] [PubMed] [Google Scholar]
- 51.Walsh G. M., Lin S., Evans D. M., Khosrovi-Eghbal A., Beavis R. C., Kast J. (2009) Implementation of a data repository-driven approach for targeted proteomics experiments by multiple reaction monitoring. J. Proteomics 72, 838–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brunner E., Ahrens C. H., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E. W., Panse C., de Lichtenberg U., Rinner O., Lee H., Pedrioli P. G., Malmstrom J., Koehler K., Schrimpf S., Krijgsveld J., Kregenow F., Hekc A. J., Hafen E., Schlapbach R., Aebersold R. (2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576–583 [DOI] [PubMed] [Google Scholar]
- 53.Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 54.Listgarten J., Emili A. (2005) Statistical and computational methods for comparative proteomic profiling using liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics 4, 419–434 [DOI] [PubMed] [Google Scholar]
- 55.Listgarten J., Neal R. M., Roweis S. T., Wong P., Emili A. (2007) Difference detection in LC-MS data for protein biomarker discovery. Bioinformatics 23, e198–e204 [DOI] [PubMed] [Google Scholar]
- 56.Radulovic D., Jelveh S., Ryu S., Hamilton T. G., Foss E., Mao Y., Emili A. (2004) Informatics platform for global proteomic profiling and biomarker discovery using liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics 3, 984–997 [DOI] [PubMed] [Google Scholar]
- 57.Gerszten R. E., Carr S. A., Sabatine M. (2010) Integration of proteomic-based tools for improved biomarkers of myocardial injury. Clin. Chem. 56, 194–201 [DOI] [PubMed] [Google Scholar]
- 58.Webb-Robertson B. J. M., Cannon W. R., Oehmen C. S., Shah A. R., Gurumoorthi V., Lipton M. S., Waters K. M. (2008) A support vector machine model for the prediction of proteotypic peptides for accurate mass and time proteomics. Bioinformatics 24, 1503–1509 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.