Abstract
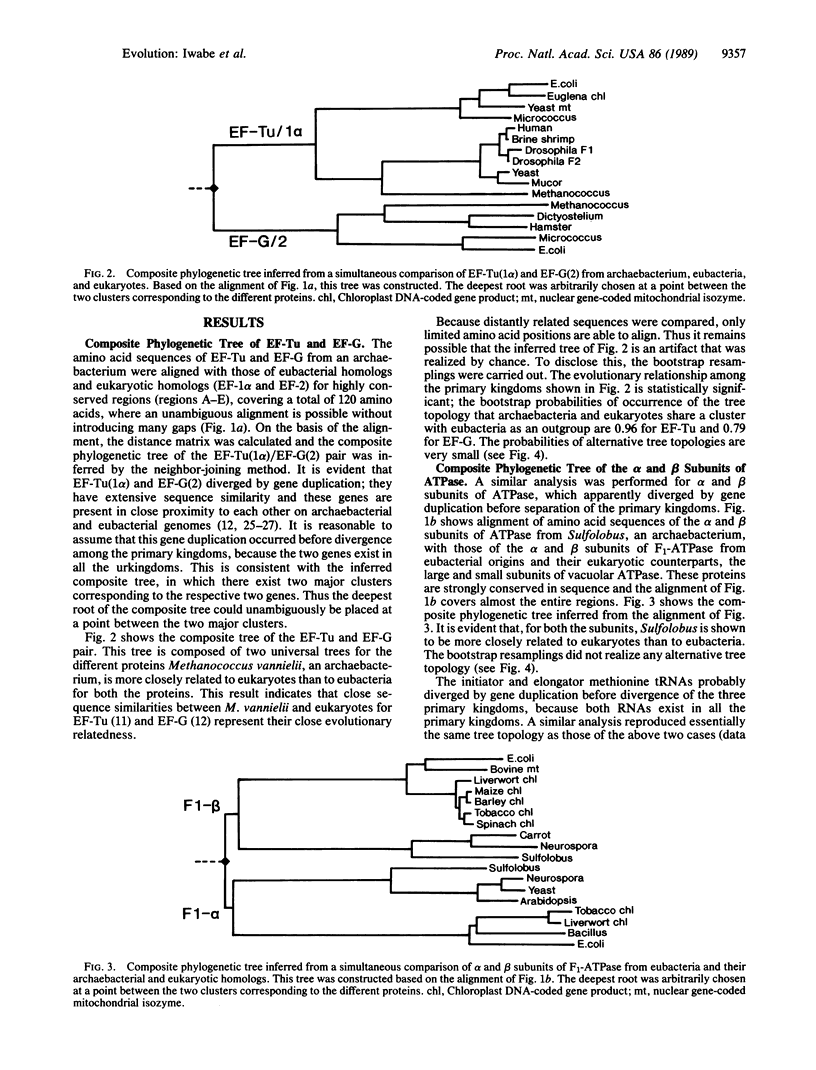
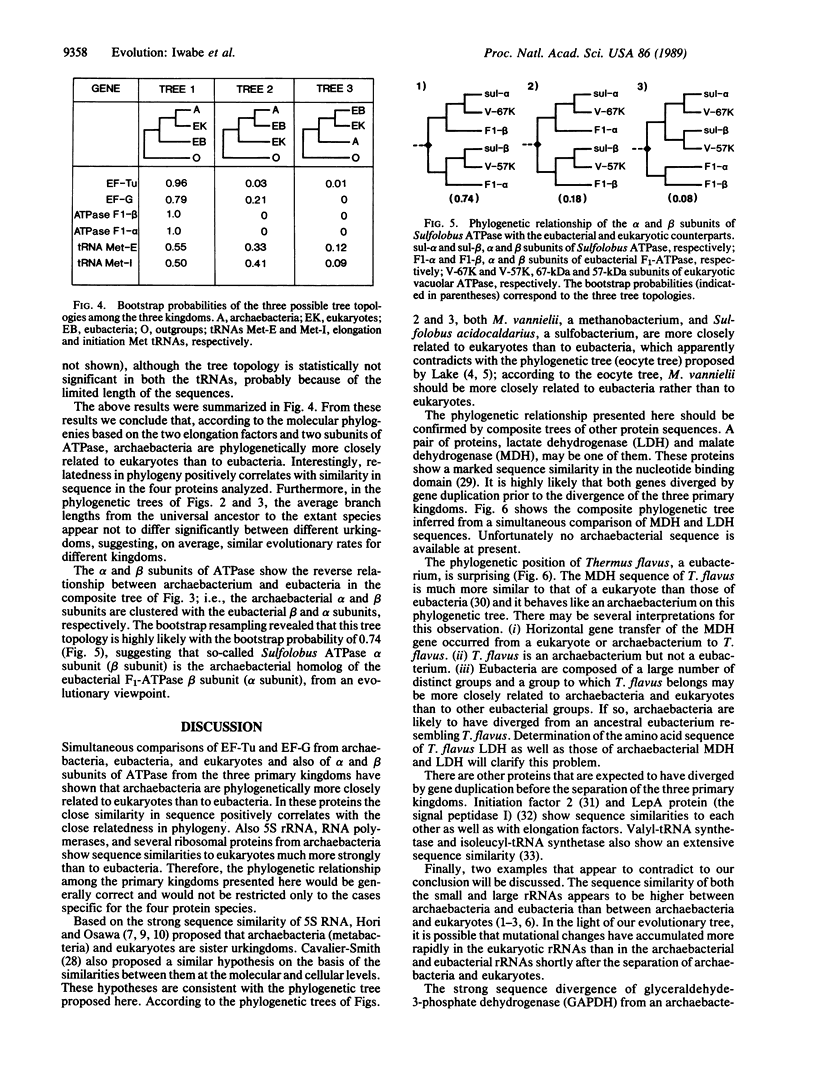
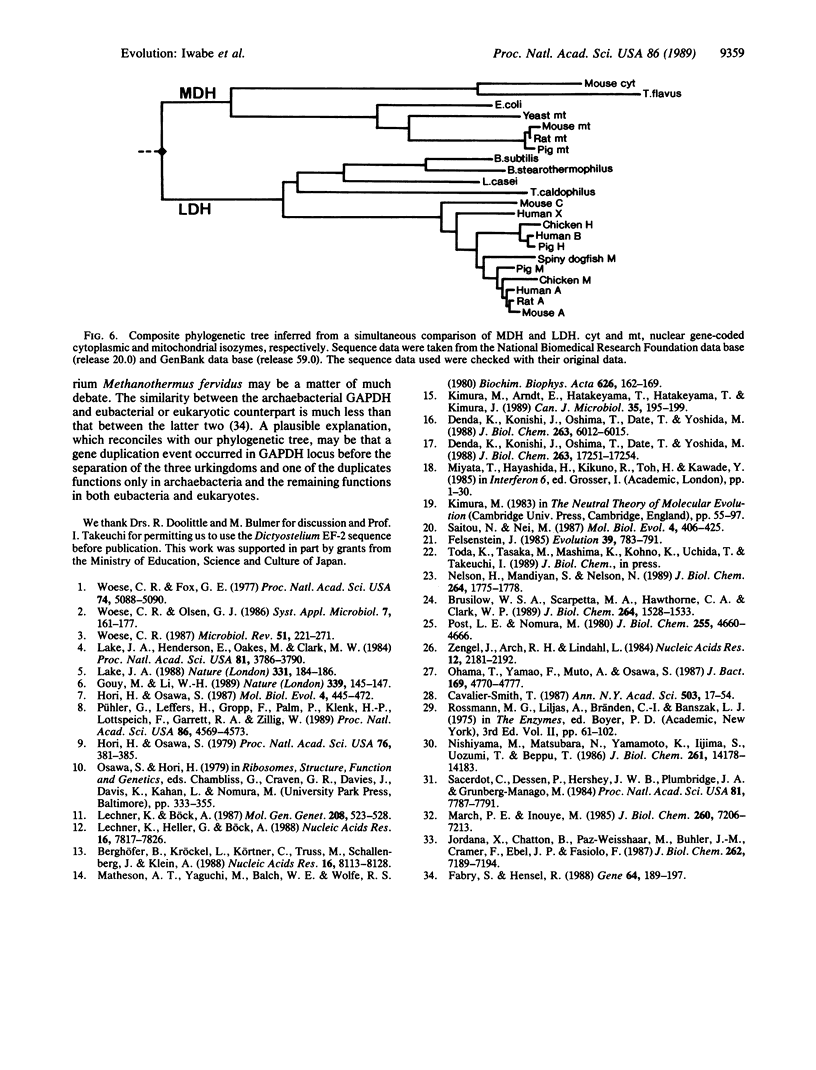
All extant organisms are though to be classified into three primary kingdoms, eubacteria, eukaryotes, and archaebacteria. The molecular evolutionary studies on the origin and evolution of archaebacteria to date have been carried out by inferring a molecular phylogenetic tree of the primary kingdoms based on comparison of a single molecule from a variety of extant species. From such comparison, it was not possible to derive the exact evolutionary relationship among the primary kingdoms, because the root of the tree could not be determined uniquely. To overcome this difficulty, we compared a pair of duplicated genes, elongation factors Tu and G, and the alpha and beta subunits of ATPase, which are thought to have diverged by gene duplication before divergence of the primary kingdoms. Using each protein pair, we inferred a composite phylogenetic tree with two clusters corresponding to different proteins, from which the evolutionary relationship of the primary kingdoms is determined uniquely. The inferred composite trees reveal that archaebacteria are more closely related to eukaryotes than to eubacteria for all the cases. By bootstrap resamplings, this relationship is reproduced with probabilities of 0.96, 0.79, 1.0, and 1.0 for elongation factors Tu and G and for ATPase subunits alpha and beta, respectively. There are also several lines of evidence for the close sequence similarity between archaebacteria and eukaryotes. Thus we propose that this tree topology represents the general evolutionary relationship among the three primary kingdoms.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berghöfer B., Kröckel L., Körtner C., Truss M., Schallenberg J., Klein A. Relatedness of archaebacterial RNA polymerase core subunits to their eubacterial and eukaryotic equivalents. Nucleic Acids Res. 1988 Aug 25;16(16):8113–8128. doi: 10.1093/nar/16.16.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusilow W. S., Scarpetta M. A., Hawthorne C. A., Clark W. P. Organization and sequence of the genes coding for the proton-translocating ATPase of Bacillus megaterium. J Biol Chem. 1989 Jan 25;264(3):1528–1533. [PubMed] [Google Scholar]
- Cavalier-Smith T. The origin of eukaryotic and archaebacterial cells. Ann N Y Acad Sci. 1987;503:17–54. doi: 10.1111/j.1749-6632.1987.tb40596.x. [DOI] [PubMed] [Google Scholar]
- Denda K., Konishi J., Oshima T., Date T., Yoshida M. Molecular cloning of the beta-subunit of a possible non-F0F1 type ATP synthase from the acidothermophilic archaebacterium, Sulfolobus acidocaldarius. J Biol Chem. 1988 Nov 25;263(33):17251–17254. [PubMed] [Google Scholar]
- Denda K., Konishi J., Oshima T., Date T., Yoshida M. The membrane-associated ATPase from Sulfolobus acidocaldarius is distantly related to F1-ATPase as assessed from the primary structure of its alpha-subunit. J Biol Chem. 1988 May 5;263(13):6012–6015. [PubMed] [Google Scholar]
- Fabry S., Hensel R. Primary structure of glyceraldehyde-3-phosphate dehydrogenase deduced from the nucleotide sequence of the thermophilic archaebacterium Methanothermus fervidus. Gene. 1988 Apr 29;64(2):189–197. doi: 10.1016/0378-1119(88)90334-4. [DOI] [PubMed] [Google Scholar]
- Gouy M., Li W. H. Phylogenetic analysis based on rRNA sequences supports the archaebacterial rather than the eocyte tree. Nature. 1989 May 11;339(6220):145–147. doi: 10.1038/339145a0. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Osawa S. Origin and evolution of organisms as deduced from 5S ribosomal RNA sequences. Mol Biol Evol. 1987 Sep;4(5):445–472. doi: 10.1093/oxfordjournals.molbev.a040455. [DOI] [PubMed] [Google Scholar]
- Jordana X., Chatton B., Paz-Weisshaar M., Buhler J. M., Cramer F., Ebel J. P., Fasiolo F. Structure of the yeast valyl-tRNA synthetase gene (VASI) and the homology of its translated amino acid sequence with Escherichia coli isoleucyl-tRNA synthetase. J Biol Chem. 1987 May 25;262(15):7189–7194. [PubMed] [Google Scholar]
- Kimura M., Arndt E., Hatakeyama T., Hatakeyama T., Kimura J. Ribosomal proteins in halobacteria. Can J Microbiol. 1989 Jan;35(1):195–199. doi: 10.1139/m89-030. [DOI] [PubMed] [Google Scholar]
- Lake J. A., Henderson E., Oakes M., Clark M. W. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3786–3790. doi: 10.1073/pnas.81.12.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 1988 Jan 14;331(6152):184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- Lechner K., Heller G., Böck A. Gene for the diphtheria toxin-susceptible elongation factor 2 from Methanococcus vannielii. Nucleic Acids Res. 1988 Aug 25;16(16):7817–7826. doi: 10.1093/nar/16.16.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March P. E., Inouye M. Characterization of the lep operon of Escherichia coli. Identification of the promoter and the gene upstream of the signal peptidase I gene. J Biol Chem. 1985 Jun 25;260(12):7206–7213. [PubMed] [Google Scholar]
- Matheson A. T., Yaguchi M., Balch W. E., Wolfe R. S. Sequence homologies in the N-terminal region of the ribosomal 'A' proteins from Methanobacterium Thermoautotrophicum and Halobacterium cutirubrum. Biochim Biophys Acta. 1980 Nov 20;626(1):162–169. doi: 10.1016/0005-2795(80)90207-x. [DOI] [PubMed] [Google Scholar]
- Miyata T., Hayashida H., Kikuno R., Toh H., Kawade Y. Evolution of interferon genes. Interferon. 1985;6:1–30. [PubMed] [Google Scholar]
- Nelson H., Mandiyan S., Nelson N. A conserved gene encoding the 57-kDa subunit of the yeast vacuolar H+-ATPase. J Biol Chem. 1989 Jan 25;264(3):1775–1778. [PubMed] [Google Scholar]
- Nishiyama M., Matsubara N., Yamamoto K., Iijima S., Uozumi T., Beppu T. Nucleotide sequence of the malate dehydrogenase gene of Thermus flavus and its mutation directing an increase in enzyme activity. J Biol Chem. 1986 Oct 25;261(30):14178–14183. [PubMed] [Google Scholar]
- Ohama T., Yamao F., Muto A., Osawa S. Organization and codon usage of the streptomycin operon in Micrococcus luteus, a bacterium with a high genomic G + C content. J Bacteriol. 1987 Oct;169(10):4770–4777. doi: 10.1128/jb.169.10.4770-4777.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Nomura M. DNA sequences from the str operon of Escherichia coli. J Biol Chem. 1980 May 25;255(10):4660–4666. [PubMed] [Google Scholar]
- Pühler G., Leffers H., Gropp F., Palm P., Klenk H. P., Lottspeich F., Garrett R. A., Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot C., Dessen P., Hershey J. W., Plumbridge J. A., Grunberg-Manago M. Sequence of the initiation factor IF2 gene: unusual protein features and homologies with elongation factors. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7787–7791. doi: 10.1073/pnas.81.24.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Olsen G. J. Archaebacterial phylogeny: perspectives on the urkingdoms. Syst Appl Microbiol. 1986;7:161–177. doi: 10.1016/s0723-2020(86)80001-7. [DOI] [PubMed] [Google Scholar]
- Zengel J. M., Archer R. H., Lindahl L. The nucleotide sequence of the Escherichia coli fus gene, coding for elongation factor G. Nucleic Acids Res. 1984 Feb 24;12(4):2181–2192. doi: 10.1093/nar/12.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]


