Figure 1.
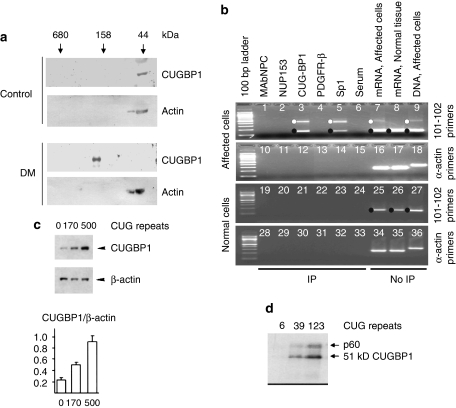
Demonstration that mut RNA binds CUGBP. (a) CUGBP in vivo is completely complexed with mut RNA. Protein from cardiac muscle was fractionated on size-exclusion chromatography and detected by antibody against CUGBP or actin (negative control). In normal hearts, CUGBP runs at 44 kDa, at its native protein MW, but in DM hearts runs at a high-MW position of 150 kDa that is RNase sensitive, corresponding to CUGn position (not shown) (from Timchenko et al8). (b) CUGBP binds only the mutant form of DMPK mRNA in vivo. Nuclear extracts underwent RT–PCR after immunoprecipitation (‘IP') or not (‘no IP'), with antibody to CUGBP, nuclear pore proteins, TF Sp1 and PDGF receptor. Anti-CUGBP coprecipitated DMPK RNA only from DM-affected, and not control, cells, using primers (101–102) spanning the CUGn repeat (n=100). Mut RNA was also precipitated with transcription factor Sp1 but not any other protein and actin RNA was not present in any IP. (Wild-type DMPK mRNA generates single band (150 nt) and mutant DMPK dual bands (150 and 450 nt), with the smaller band arising by template sliding across expanded CUGn during PCR.) (from Ebralidze et al15) (c) Binding of CUGBP to longer mut RNAs in vivo leads to increased CUGBP in cells that correlates with longer protein half-life. Upper: western blot. Lower: graph of data from three experiments (from Timchenko et al8). (d) Increased binding of CUGBP in vitro with longer repeat length. ‘RNA-free' CUGBP was mixed with equal molar quantities of end-labeled RNAs containing repeat lengths of 6, 39 and 123 CUG and assessed for binding (from Timchenko et al27).