Abstract
Hereditary recurrent fevers (HRF) are rare diseases caused by molecular defects in genes involved in the regulation of innate immunity. Sixty-seven international laboratories participated in an external quality assessment (EQA) scheme, which was developed to appraise the accuracy of genetic testing. Reports were evaluated for the 12 items recommended by the OECD (Organisation for Economic Co-Operation and Development) guidelines for molecular diagnostics. The best documented items were the name of the gene, the biologist, or the patient, whereas information on the test and screening limits, and clinical interpretation of the disease inheritance were scarcely provided. The mutation nomenclature was incomplete in about 70% of the cases. In the first 2 years of EQA, we identified almost 30% genotyping error rate, which decreased markedly in the last year. The combined performance on the basis of the correct identification of all genotypes by a given laboratory in all the 3 years was only 40%, showing a critical need for improvement.
Keywords: EQA/PT, molecular diagnosis, hereditary recurrent fevers
Introduction
Patients suffering from hereditary recurrent fevers (HRF) (reviewed in McDermott and Frenkel1) present with unprovoked recurrent bouts of fever and serosites. Abdominal, articular, and cutaneous signs of inflammation manifest with variable intensity and localization, but may lack specificity, thus often making clinical diagnosis difficult. HRF classically include four entities: two are recessively inherited, familial Mediterranean fever ((FMF), gene MEFV, MIM 608107); and mevalonate kinase deficiency ((MKD), gene MVK, MIM 251170); and two are dominant, TNF receptor-associated periodic syndrome ((TRAPS), gene TNFRSF1A, MIM 191190); and cryopyrin-associated periodic syndrome ((CAPS), gene NLRP3, MIM 606416). A fifth one, NLRP12-associated periodic syndrome ((NAPS12), gene NLRP12, MIM 609648), was described very recently.2 HRF are rare diseases except for FMF, which is a very common disease in several Mediterranean and Middle Eastern populations. Although the transmission of these disorders matches Mendelian patterns, dominant inheritance in FMF,3 digenic inheritance,4 or incomplete penetrance5 have been described. The causative genes encode proteins involved in regulation of innate immunity, mainly through their role in the regulation of pro-inflammatory cytokines and apoptosis.6 Molecular screening of these genes is of particular interest in paucisymptomatic patients, and has dramatically improved the patient's quality of life, by providing early and accurate diagnosis and allowing subsequent appropriate treatment.
Genetic testing as a way to corroborate clinical diagnosis must comply with the rigorous quality control procedures in order to ensure consistency and accuracy of results. The evidence for the necessity to improve the quality of molecular diagnostics appeared in a surprising paper published in 2001 on genetic testing for cystic fibrosis.7 This study showed that less than half of the participating laboratories correctly identified genotypes in each of the three trials. Thirty-nine per cent made a mistake only once, whereas 2% of the laboratories consistently failed in each trial. After 5 years of experience, a clear improvement was shown. Since this initial report, additional quality control schemes have been developed for many other diseases.8, 9, 10, 11
Here, we present a 3-year trial of an external quality assessment (EQA) scheme for the molecular diagnosis of HRF, with three specific aims: to survey the spread and scope of HRF genetic testing worldwide, to evaluate the content of the reports, and to estimate the accuracy of testing.
Materials and methods
Survey of laboratories performing genetic testing for HRF
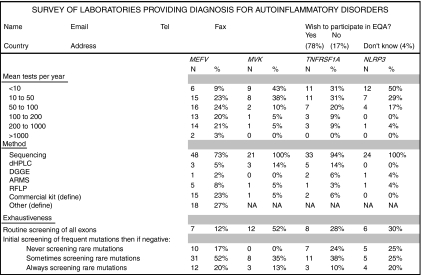
We used various sources to identify laboratories that would potentially be interested in participating in this EQA: generalist databases, Orphanet (http://www.orpha.net/consor/cgi-bin/index.php), EDDNAL (http://www.eddnal.com/), Genetests (http://www.genetests.org/), and an online registry of auto-inflammatory mutations developed in Montpellier, Infevers (http://fmf.igh.cnrs.fr/ISSAID/infevers/). We searched by the name of the disease, the gene and their aliases: familial Mediterranean fever, FMF, MEFV; mevalonate kinase deficiency, MKD, hyper-IgD syndrome, HIDS, mevalonic aciduria, MVK; TNF receptor-associated periodic syndrome, Hibernian fever, TRAPS, TNFR1, TNFRSF1A; cryopyrin-associated periodic syndrome, CAPS, familial cold autoinflammatory syndrome, FCAS, Muckle–Wells syndrome, MWS, chronic infantile neurological cutaneous and articular syndrome, CINCA, and neonatal onset multisystem inflammatory disease, NOMID, CIAS1, NALP3, PYPAF1, cryopyrin, NLRP3. One hundred and forty laboratories were retrieved and contacted by email. Thirty-one additional laboratories heard independently about the quality control network project and contacted us spontaneously during the framework of this 3-year scheme. They all received a questionnaire about their activities (Figure 1).
Figure 1.
Questionnaire on the laboratories activity and results of the survey. The empty questionnaire was sent by email as an attached excel file to the laboratories identified through various web sources. Results obtained after the survey are shown for each item. For the screening methods, the sum exceeds 100% as several laboratories combined various molecular approaches. The commercial kits used for FMF testing were the FMF StripAssay (ViennaLab Diagnostics GmbH, Vienna, Austria), N=17 and the PRONTO FMF Screen™, Pronto Diagnostics Ltd, Rehovot, Israel), N=1. MEFV: Mediterranean fever, MVK: mevalonate kinase, TNFRSF1A: TNF receptor superfamily member 1A, NLRP3: NLR family, pyrin domain containing 3, DHPLC: denaturing high performance liquid chromatography, DGGE: denaturing gel gradient electrophoresis, ARMS: amplification refractory mutation system, RFLP: restriction fragment length polymorphisms.
Organization of the EQA
The EQA scheme described in this study ran from 2006 to 2008 and, to our knowledge, is the only such initiative done for genetic testing of HRF. The laboratories that participated in the EQA were allocated a unique ID number. They were requested to contribute anonymous DNA samples to constitute a repository of mutant DNA samples. The samples were received in the coordinating centre (I Touitou, Montpellier) and were all checked for their quality (by measuring the optical density at 260 and 280 nm), and for genotypes (by sequencing) before being sent to the participants. Depending on the number of the HRF genes being tested in a given laboratory, 1–4 DNA samples (1 sample per gene and per year) were sent to each participant. The mutations expected to be identified in the test samples in each year of EQA are shown in Table 1. An effort was taken to assemble a balanced number of common and unusual genotypes. Clinical information was not provided to mimic usual laboratory situations. Genetic reports were requested to be issued in English and sent by email to the coordinator. Although most participating laboratories were not native in English, we believe that this request was unlikely to affect the way they routinely proceed for both molecular analysis and reporting.
Table 1. Genotypes of hereditary recurrent fevers distributed in the 3-year molecular diagnosis EQA scheme, and wrongly reported genotypes.
| Expected names | |||||||
|---|---|---|---|---|---|---|---|
| Year | Gene | HGVS DNA name | HGVS protein name | Usual namea | List of incorrect reported genotypes | ||
| 2006 | MEFV | c.442G>C;c.2076_2078del | p.Glu148Gln;p.Ile692del | E148Q;I692del | E148Q+M694del | S747S | |
| MVK | c.1129G>A | p.Val377Ile | V377I | ||||
| TNFRSF1A | c.362G>A | p.Arg121Gln | R92Q | ||||
| NLRP3 | No mutation | No mutation | No mutation | Q703K (2 times) | |||
| 2007b | MEFV | NM_000243.1:c.[2080A>G(+) 2230G>T] | p.[Met694Val(+)Ala744Ser] | M694V+A744S | M694V only | ||
| MEFV | NM_000243.1:c.896A>G | p.[Glu299Gly]+[?] | E299G | No mutation (two times) | E148Q | 896C>G | |
| MVK | NM_000431.1:c.[803T> C(+)1129G>A] | p.[Ile268Thr(+)Val377Ile] | I268T+V377I | V377I only | I268T only | ||
| TNFRSF1A | NM_001065.2:c.204G>C | p.Leu68Phe | L39F | No mutation | P12P | C39F | |
| NLRP3 | NM_004895.3:c.1706G>C | p.Gly569Ala | G569A | G569R (three times) | G571A | 1707G>C | |
| 2008 | MEFV | NM_000243.1:c.[2040G>C(+) 2080A>G] | p.[Met680Ile(+)Met694Val] | M680I+M694V | |||
| MVK | NM_000431.1:c.[1039+1G> T(+)1129G>A] | p.[Glu296GlyfsX14(+) Val377Ile] | E296Gfs+V377I | V377I only | |||
| TNFRSF1A | NM_001065.2:c.204G>C | p.Pro75Leu | P46L | No mutation | |||
| NLRP3 | NM_004895.3:c.907G>A | p.Asp303Asn | D303N | ||||
HGVS: Human Genome Variation Society. The HGVS names refer to the official nomenclature at: http://www.hgvs.org/mutnomen/.
The usual name refers to the name as initially published or submitted to Infevers, the registry of auto-inflammatory mutations at http://fmf.igh.cnrs.fr/ISSAID/infevers/.
Two different MEFV samples were used for the 2007 round. They were randomly assigned to the participants.
Evaluation of the report contents
Criteria for the report content and mutation nomenclature were not pre-defined as our goal was to overview the current practices. We used the OECD (Organisation for Economic Co-Operation and Development) guidelines for quality assurance in molecular genetic testing (http://www.oecd.org/dataoecd/43/6/38839788.pdf). We evaluated the reporting practices and rated the 12 minimal items that should be found in reports as recommended by the OECD, but we did not check whether laboratories were accredited, certified, or used qualified personnel. The mutation nomenclature as recommended by the Human Genome Variation Society (http://www.hgvs.org/mutnomen) was the reference for testing performance.12, 13 A genotype was rated as correct in relation to the technique used, even if the nomenclature was not complete. To facilitate analysis of the reports, we constructed a database (Microsoft Access) containing fields for the following items: the details on the laboratories, the genotype found in the samples, and the conclusions given by the laboratories. Anonymized detailed reports, the EQA statistics, and a certificate of participation were sent to all participants after each year of EQA scheme.
Results
HRF genetic testing survey and EQA participants
We identified 171 laboratories worldwide that perform genetic testing for HRF. Of these, 83 laboratories replied over the 3 years to the questionnaire detailing the scope and methods of their genetic testing activities. To the question whether they would agree to participate in an EQA scheme for molecular diagnosis of HRF, 65 laboratories (78%) responded ‘yes', 14 (17%) responded ‘no', and the 4 remaining responded ‘unknown'. Two of the latter finally participated. More than half (52%) of the laboratories tested only for the most common disease, FMF (MEFV gene), whereas only 18% of laboratories provided molecular diagnostics for all four HRF. The number of samples analyzed per laboratory ranged from <10 to over 1000 per year, with MEFV testing reaching over 200 samples per year in 24% of the participating laboratories (Figure 1). The most common molecular technique used for genetic testing was sequencing of mutational hot spots (81% of the laboratories), whereas 40% of the laboratories used alternative or additional screening methods (denaturing high-performance liquid chromatography, denaturing gel gradient electrophoresis), or mutation-specific screening approaches (amplification refractory mutation system, restriction fragment length polymorphisms). As expected, only a small number of laboratories routinely screen for mutations in all exons of a given gene with the exception of MVK. This is likely because of the fact that MVK disease-associated mutations are spread throughout the gene.
Report issuing and content
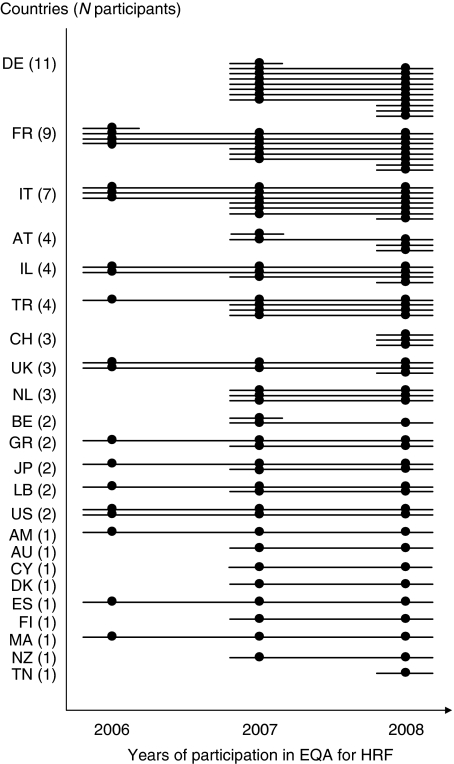
A total of 23 countries joined the project, mostly from Europe but also from the United States, New Zealand, Japan, and Australia (Figure 2 and the list of participants in acknowledgements). We observed an under-representation of countries where FMF is very prevalent, yet a nice representation of emigration countries with large emigrations from Mediterranean countries, for example, Germany and Austria with Turkish population, France with Magreb Arabs.
Figure 2.
World distribution of the participating laboratories. The lines illustrate the number of centers active in the EQA scheme in each country. The circles indicate the year(s) of participation for a given centre. The dropout rate (1/23 and 3/52 participants centers in 2006 and 2007, respectively) was about 5%.
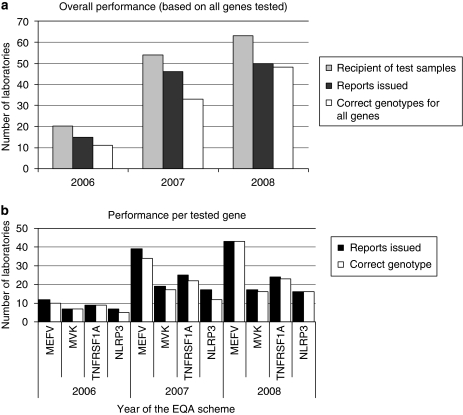
The number of laboratories who expressed their willingness to receive EQA samples tripled over the 3 years (20 in 2006, 63 in 2008, Figures 2 and 3a). Eighty per cent of them issued genetic reports for the EQA samples over the 3 years. The mean time for reporting was <2 months and ranged from 1 day (in case of use of the commercial kit for the MEFV testing) to over 5 months. There was a slight trend for faster reporting by laboratories that process large number of samples per year, but this was not significant (data not shown).
Figure 3.
Analysis of performance (participation, reporting ,and correct genotyping) in the EQA scheme per laboratory and per gene. (a) The number of laboratories that expressed their willingness to participate in each EQA scheme, and received samples is represented in gray. The number of laboratories that issued reports and gave correct mutations for the genes tested is indicated in black and white, respectively. (b) Results are expressed according to the genes tested.
The reports varied in length and content. For instance, five participants sent the results as a crude genotype in the core of an email, whereas one sent a report consisting of four attached pages per tested gene, with details including polymorphisms and references. The OECD-recommended items were checked yearly once for each participating laboratory, regardless of the report format. Table 2 shows the detailed statistics of this study. The best documented items in the laboratory reports were the patient name (94–100%), the name of the gene (87–98%), the laboratory address (72–94%), and the referring physician (74–80%); however, even this key information was absent in a significant number of reports. Items such as information on the detection rate (20–26%) or interpretation of the testing results according to the mode of inheritance of the disease (8–47%) were scarcely provided. The mutation nomenclature as recommended by the HGVS was complete in less than one-third of the cases but not necessarily wrong.
Table 2. Rate of compliance (%) with the recommended OECD items over years.
| Item | 2006 | 2007 | 2008 |
|---|---|---|---|
| 1. Information linking the report to the patient | |||
| Name of the patient | 100.0% | 93.5% | 98.0% |
| Sample # | 60.0% | 63.0% | 68.0% |
| Date of birth of the patient | 53.3% | 50.0% | 60.0% |
| 2. Name/address of the referring physician | 80.0% | 73.9% | 78.0% |
| 3. Indication for the test (disease name) | 100.0% | 82.6% | 84.0% |
| 4. Details on the test performed | |||
| Gene tested | 86.7% | 97.8% | 96.0% |
| Mutation(s) tested | 86.7% | 93.5% | 84.0% |
| Methods used | 86.7% | 84.8% | 86.0% |
| Detection rate (screening exhaustivenessa) | 20.0% | 26.1% | 24.0% |
| Error risk (test limits) | 13.3% | 8.7% | 18.0% |
| 5. Nature of sample (DNA or blood) | 60.0% | 78.3% | 70.0% |
| 6. Date of sample reception | 33.3% | 65.2% | 65.2% |
| 7 (and 11.). Laboratory details | 93.3% | 93.5% | 72.0% |
| 8. Results | |||
| Genotype(s) correctb | 73.0% | 71.7% | 96.0% |
| Nomenclature complete | 33.3% | 23.9% | 28.0% |
| 9. Interpretation | |||
| Comments (disease confirmation?) | Not rated | 78.3% | 84.0% |
| Cis or trans? (recessive diseases) | 46.7% | 36.0% | 41.9% |
| De novo? (dominant disease) | Not relevant | 8.0% | 17.4% |
| Further explanation/genetic counselling | 26.7% | 39.1% | 50.0% |
| 10. Name of the biologist | 93.3% | 95.7% | 100.0% |
| 12. Date of report | 93.3% | 80.4% | 96.0% |
Items with satisfying responses are in bold letters.
As described in Figure 1.
Rated as correct if all of the genes tested were rightly reported by the concerned participant.
Genotyping performance and interpretation issues
The more concerning issue was the high rate of genotyping errors (Table 1). Mutations were incorrect at the nucleotide or the protein levels or both, whereas some were likely mistyped. A trend toward a better performance was observed in laboratories performing a large number of analyses per year, but this was not significant (data not shown). Figure 3b describes the rate of correct genotyping per year and per gene. The worst performance was seen for the NLRP3 gene, possibly because it is a recently discovered gene and most patients have private mutations. Almost 30% of the laboratories made at least one genotyping error between 2006 and 2007 (Figure 3a). During the 2006 EQA, one laboratory reported wrong genotypes for three out of the four tested genes. At the combined performance, only 40% of the laboratories that participated in all the 3 years of EQA made no genotyping errors for all of the genes they tested.
Several issues related to the interpretation of the genotypes are likely to account for this poor performance. In the first year of EQA, the MEFV test sample (Table 1) contained the p.E148Q mutation and the p.I692del deletion. These two variants often co-segregate in the form of a complex allele. No information was provided to the participants on the parental genotypes in order to evaluate their interpretation of this genotype. Actually, the test sample was a carrier for a complex allele with no mutation on the second chromosome. On the basis of the sole finding of two mutations, two laboratories concluded that the diagnosis of recessively inherited FMF was genetically confirmed. Another participant reported mutation of p.I694del instead of p.I692del.
In the second year of EQA, one sample contained the p.E299G substitution, a rare mutation in MEFV exon 2 that is not explored by any of the available kits for mutation detection in the MEFV gene. A genetic report lacking this mutation was accepted as correct provided that it commented about the limited exhaustiveness of the method used for mutation screening, and about the possibility that rare mutations are not investigated by their screening strategy. The NLRP3 mutant EQA sample harbored a novel p.G569A mutation. However, as a similar mutation at the same residue, p.G569R, was recorded in Infevers, three laboratories mistakenly reported the p.G569R mutation instead of p.G569A, giving Infevers as the reference.
In the third round of EQA, the MVK sample had a splice mutation that was not identified by two laboratories. The result was accepted as correct for one of them, whose report included an explanation about limitations of the screening strategy. The TNFRSF1A mutant sample carried the p.P75L (usual name P46L) variant, which is found at a low frequency in the Arabian and African control populations. This variant is often found in patients with TRAPS-like disease and functional studies supported its role in inflammation. As the functional significance of p.P75L is still under debate, only reports with a statement that this variant is of unknown clinical significance were accepted as correct, and a report with a statement that ‘no pathogenic mutations' were identified in the sample was rated as incorrect.
Discussion
The recent discovery of several causative genes for HRF has prompted a large number of laboratories to develop a genetic diagnosis for these diseases. However, very little was known about the availability of these new tests worldwide, the panel of genes analyzed, the form and content of genetic reports, or proficiency testing. To assess these issues, we developed a ‘home-made' international EQA scheme, which was preceded by a laboratory survey. Although HRF are rare diseases, we identified an unexpected 171 laboratories providing molecular diagnosis for one to four of these disorders. A close survey of 83 of these laboratories shows that genetic testing of FMF, the most prevalent HRF, is now broadly available, but very few laboratories (18%) offer molecular testing for MKD, TRAPS, and CAPS genes. However, we found that the FMF genetic testing from many countries with a high prevalence of the disease was under-represented in our EQA scheme. In contrast, FMF diagnostic centers from their prospective emigration countries comprised the bulk of the study group (Figure 2). The fact that there may be a deficiency in test availability in high-risk countries brings up health-policy issues that need to be addressed in the future.
This study highlighted an extreme heterogeneity in the depth and format of laboratory reports. The minimal information content for the prescribing practitioner, as recommended by the OECD guidelines for molecular testing, was most often lacking, indicating that these guidelines are slow to be adopted in practice. This study also showed that the main issues to be addressed and improved in genetic testing of HRF were the accuracy of genotyping and mutation nomenclature. Indeed, the genotypes delivered during the first two rounds of EQA were wrong in one-third of the cases, showing a significant need for continuous monitoring and improvement in the quality of results. Although the performance for HRF testing in the first 2 years of EQA was as poor as in the initial BRCA scheme, only 4% of the participants failed to report correct genotypes in the last EQA round. There are several possible explanations for the observed improvement. The compiled results of the first EQA years were reported to the participating laboratories in an anonymous but identifiable mode, prompting some laboratories to be more alert in genotype identification and reporting during the last year of EQA. The second possible explanation is that in the last round of EQA we distributed DNA samples carrying more common and thus more familiar HRF mutations.
Our scheme has been recently referenced by the EuroGentest European Quality Assurance Database (http://www.eurogentest.org/web/qa/basic.xhtml). We plan to compose a standardized genetic report for testing of HRF and make it available to all participants. We also plan to join the European Molecular Genetics Quality Network (http://www.emqn.org/emqn/), and to establish guidelines for molecular diagnosis and reporting. Indeed, harmonization of genetic testing procedures, and assurance that all laboratories meet the minimum levels of expertise and follow standardized protocols is critical in the field of rare diseases. This is a timely step in the face of the increase in the awareness, geographical spread, and density of HFR diagnostic services.
Acknowledgments
This work was supported by the French Ministry of Health and the Centre Hospitalo-Universitaire de Montpellier and through Coordination Theme 1 (Health) of the European Community's FP7. We thank Dr J Alexander for editing the paper. We are grateful to Dr P Taschener and Dr J den Dunnen for their help in setting correct nomenclature for HRF mutations as recommended by the HGVS society at http://www.hgvs.org/mutnomen/. We thank all the colleagues who played the game and participated in one or more of these schemes: Dr Akar (TR), Dr Aksentijevich (US), Dr Altiok (TR), Dr Austrup (DE), Dr Bakker (NL), Dr Baldi (IT), Dr Belmahi (MA), Dr Bennetts (AU), Dr Bourmeyster (FR), Dr Burwinkel (DE), Dr Bybee (GB), Dr Ceccherini (IT), Dr Chaabouni (TN), Dr Costa (FR), Dr Costi (CY), Dr Cuisset (FR), Dr Dode (FR), Dr Dutly (CH), Dr Etlik (TR), Dr Franke (DE), Dr Gershoni-Baruch (IL), Dr Goulielmos (GR), Dr Haverkamp (DE), Dr Hilbert (BE), Dr Hujeirat (IL), Dr Ida (JP), Dr Konstantopoulos (GR), Dr Legendre (FR), Dr Lerer (IL), Dr Lohse (DE), Dr Mahfouz (LB), Dr Maiwald (DE), Dr Marschall (DE), Dr Martorana (IT), Dr Mcdermott (GB), Dr Medlej-Hashim (LB), Dr Moritz (AT), Dr Morris (CH), Dr Neri (IT), Dr Nevinny-Stickel-Hinzpeter (DE), Dr Nishikomori (JP), Dr Oberkanins (AT), Dr Obici (IT), Dr Parma (BE), Dr Patrosso (IT), Dr Pingault (FR), Dr Pontillo (IT), Dr Potter (NZ), Dr Renner (AT), Dr Richard (US), Dr Rogge (DE), Dr Ronsin (FR), Dr Sarkisian (AM), Dr Schwartz (DK), Dr Shinar (IL), Dr Sinke (NL), Dr Skladny (DE), Dr Stjernberg-Salmela (FI), Dr Tchernitchko (FR), Dr Thonney (CH), Dr Tighe (GB), Dr Touitou (FR), Dr Überlacker (DE), Dr Waterham (NL), Dr Wildhardt (DE), Dr Yagüe Ribes (ES), Dr Yilmaz (TR), and Dr Zeitlhofer (AT).
References
- McDermott MF, Frenkel J. Hereditary periodic fever syndromes. Neth J Med. 2001;59:118–125. doi: 10.1016/s0300-2977(01)00149-8. [DOI] [PubMed] [Google Scholar]
- Jeru I, Duquesnoy P, Fernandes-Alnemri T, et al. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci USA. 2008;105:1614–1619. doi: 10.1073/pnas.0708616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DR, Gillmore JD, Lachmann HJ, et al. The genetic basis of autosomal dominant familial Mediterranean fever. Q J Med. 2000;93:217–221. doi: 10.1093/qjmed/93.4.217. [DOI] [PubMed] [Google Scholar]
- Touitou I, Perez C, Dumont B, Federici L, Jorgensen C. Refractory auto-inflammatory syndrome associated with digenic transmission of low-penetrance tumour necrosis factor receptor-associated periodic syndrome and cryopyrin-associated periodic syndrome mutations. Ann Rheum Dis. 2006;65:1530–1531. doi: 10.1136/ard.2006.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksentijevich I, Torosyan Y, Samuels J, et al. Mutation and haplotype studies of familial Mediterranean fever reveal new ancestral relationships and evidence for a high carrier frequency with reduced penetrance in the Ashkenazi Jewish population. Am J Hum Genet. 1999;64:949–962. doi: 10.1086/302327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A, van der Meer JW. Pathogenesis of familial periodic fever syndromes or hereditary autoinflammatory syndromes. Am J Physiol Regul Integr Comp Physiol. 2007;292:R86–R98. doi: 10.1152/ajpregu.00504.2006. [DOI] [PubMed] [Google Scholar]
- Dequeker E, Ramsden S, Grody WW, Stenzel TT, Barton DE. Quality control in molecular genetic testing. Nat Rev Genet. 2001;2:717–723. doi: 10.1038/35088588. [DOI] [PubMed] [Google Scholar]
- Jennings I, Kitchen S, Woods TA, Preston FE. Multilaboratory testing in thrombophilia through the United Kingdom National External Quality Assessment Scheme (Blood Coagulation) Quality Assurance Program. Semin Thromb Hemost. 2005;31:66–72. doi: 10.1055/s-2005-863807. [DOI] [PubMed] [Google Scholar]
- Patton SJ, Wallace AJ, Elles R. Benchmark for evaluating the quality of DNA sequencing: proposal from an international external quality assessment scheme. Clin Chem. 2006;52:728–736. doi: 10.1373/clinchem.2005.061887. [DOI] [PubMed] [Google Scholar]
- Ramsden SC, Daly S, Geilenkeuser WJ, et al. EQUAL-quant: an international external quality assessment scheme for real-time PCR. Clin Chem. 2006;52:1584–1591. doi: 10.1373/clinchem.2005.066019. [DOI] [PubMed] [Google Scholar]
- Seneca S, Morris MA, Patton S, Elles R, Sequeiros J. Experience and outcome of 3 years of a European EQA scheme for genetic testing of the spinocerebellar ataxias. Eur J Hum Genet. 2008;16:913–920. doi: 10.1038/ejhg.2008.29. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Paalman MH. Standardizing mutation nomenclature: why bother. Hum Mutat. 2003;22:181–182. doi: 10.1002/humu.10262. [DOI] [PubMed] [Google Scholar]