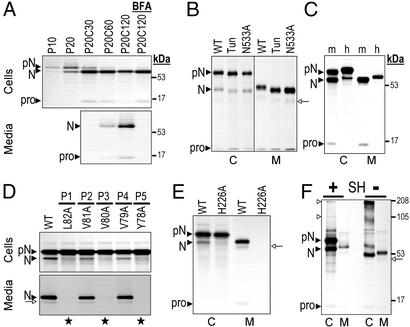
Figure 2.
Biosynthesis of NARC-1 and its mutants. CHO (A) or HK293 (B–F) cells stably or transiently transfected, respectively, with vectors expressing hNARC-1-V5 (A–F) or mNARC-1-V5 (C) were pulse-labeled (P) with [35S]EasyTag Express mix for the indicated time (in min), 4 h (B–E), or 2 h (F). Cell extracts (C) and media (M) were immunoprecipitated with a V5 antibody and the precipitates were resolved by SDS/PAGE on an 8% tricine (A and C–F) or glycine (B) gel. The migration positions of molecular mass standards (kDa), pro-NARC-1 (pN), NARC-1 (N), its secondary site-cleaved product (open arrow), or its pro-segment (pro) are emphasized. (A) Pulse–chase analysis in the absence or presence of brefeldin A (BFA). (B) Effect of tunicamycin (Tun) and the N-glycosylation mutant N533A on NARC-1 processing. (C) Comparison of mNARC-1 and hNARC-1 processing. (D) Processing of WT, P1, P2, P3, P4, and P5 Ala mutants; stars indicate the absence of processing. (E) Processing of the active site mutant H226A. (F) Oligomerization (open triangles) of proNARC-1 in the presence (+) or absence (−) of reducing agent (SH).