Abstract
The Saami from Fennoscandia are believed to represent an ancient, genetically isolated population with no evidence of population expansion. Theoretical work has indicated that under this demographic scenario, extensive linkage disequilibrium (LD) is generated by genetic drift. Therefore, it has been suggested that the Saami would be particularly suited for genetic association studies, offering a substantial power advantage and allowing more economic study designs. However, no study has yet assessed this claim. As part of a GWAS for a complex trait, we evaluated the relative power for association studies of common variants in the Finnish Saami. LD patterns in the Saami were very similar to those in the non-African HapMap reference panels. Haplotype diversity was reduced and, on average, levels of LD were higher in the Saami as compared with those in the HapMap panels. However, using a ‘hidden' SNP approach we show that this does not translate into a power gain in association studies. Contrary to earlier claims, we show that for a given set of common SNPs, genomic coverage attained in the Saami is similar to that in the non-African HapMap panels. Nevertheless, the reduced haplotype diversity could potentially facilitate gene identification, especially if multiple rare variants play a role in disease etiology. Our results further indicate that the HapMap is a useful resource for genetic studies in the Saami.
Keywords: Saami, genome-wide association studies, linkage disequilibrium, population isolates
Introduction
Population isolates have proved very useful for the identification of genes for rare Mendelian diseases.1 Their utility in elucidating the genetic basis of complex diseases, however, has been a matter of much debate in the past.2, 3, 4, 5 This uncertainty has now largely been resolved. During the last few years, numerous variants associated with complex diseases and traits have been identified in population isolates.6 One important advantage some of these populations offer, is that they show substantially higher levels of linkage disequilibrium (LD) and fewer regions of very low LD compared with outbred populations.7, 8 This makes them particularly suited for genome-wide association studies (GWASs), which rely on the LD between the typed markers and the untyped causative variants. LD that stretches out over longer distances, leads to a better genomic coverage for a given marker density. Equivalently, isolates would allow a more economic study design, in which a high genomic coverage can be attained using relatively fewer markers. It has been suggested that, in some isolates, an association study would require at least 30% fewer markers than a study in an outbred population.8
To date, gene mapping efforts in population isolates have focused on populations that were founded by a small number of individuals and that have subsequently undergone rapid exponential growth and low immigration. Recent founder populations in particular, exhibit substantially increased levels of LD as compared with outbred populations.7, 8 Examples of such young isolates can be found in Finland, the Central Valley of Costa Rica and on Sardinia.
Theoretical work has shown that a completely different demographic history can also lead to elevated levels of LD, in the absence of a founder effect. In small populations that remain constant in size over long time periods, new LD is generated by genetic drift.9, 10 This has led to the proposal of ‘drift mapping' as a gene discovery strategy.10 Chromosomal regions implicated in a complex phenotype would then be identified in a small, ancient population of stable size, using a map of modest marker density. This initial coarse mapping would subsequently be followed by fine-scale localization in an outbred population using a denser map. Isolates with this demographic past have, however, been largely ignored by geneticists due to their small population size and low recessive disease prevalence.5 Consequently, elaborate genome-wide, SNP-based studies of the magnitude and distribution of LD in such populations have not been performed.
The Saami of northern Scandinavia and the Kola Peninsula exemplify an ancient population with no evidence of expansion.2, 11, 12, 13 Among European populations, the Saami are considered a genetic ‘outlier' because of the relatively high genetic differentiation between them and other European populations, including their geographic and linguistic neighbors, the Finns.12, 14 Due to their demographic history, it has been suggested that the Saami offer great potential for ‘drift mapping', and hence, more economic GWASs.2, 10, 13, 15, 16, 17 However, this has been substantiated with only very limited empirical data, most of which predate the HapMap project.
In this paper, we present the results of a first genome-wide, SNP-based survey of the extent and distribution of LD and haplotype diversity in the Finnish Saami. We compare relative power for association studies of common SNPs with that in the HapMap reference panels, and discuss the implications for GWASs for complex phenotypes.
Materials and methods
Data sets
This study was conducted within the framework of the European ARHI project (QLRT-2001-00331) that aimed to identify environmental and genetic risk factors for age-related hearing impairment.18, 19, 20 Due to the anticipated statistical power advantage for association-based gene mapping, we conducted a GWAS in the Finnish Saami. The results of this association scan will be published elsewhere. Here we describe the results of our evaluation of relative genomic coverage.
Blood samples from Saami subjects, aged between 50 and 75 years, were collected across the north of Finland. Since this was a quantitative trait association study, there was no ascertainment based on phenotype. The eligible subjects were recruited with the aid of the public population register through a three-stage process. In a first stage, a geographical criterion was applied: only areas with a high probability of Saami inhabitation, were considered. In a second stage, putative study participants were invited based on an evaluation of Saami communities made by an expert. Finally, Saami identity of the subject in question was confirmed in a direct interview with the subject. Written informed consent was obtained from all study participants and all the samples were anonymized, with no identification of individual subjects possible. This study has been approved by the Finnish National Advisory Board on Health Care Ethics, and by the ethics committees or the appropriate local institutional review boards at all participating institutions.
Genomic DNA from 352 subjects in total was extracted from blood and diluted to 50 ng/μl. Each sample was genotyped on the Affymetrix GeneChip 100K array pair (116 204 SNPs). Genotype calling was performed using the BRLMM algorithm in the Affymetrix GeneChip Genotyping Analysis Software (GTYPE) version 4.1. Data management and quality control were performed using the PLINK toolset21 (http://pngu.mgh.harvard.edu/purcell/plink/). Eight subjects were removed due to either a low sample call rate (<94%), an unintentional sample duplication event or a sample switch event. The average sample call rate in the remaining 344 subjects was 99.2%.
To evaluate the magnitude of LD, haplotype diversity and power for genetic association studies, we obtained the genotype data of the International HapMap Project (phase 2; release 23).22, 23 This data set contains information on 3.96 million SNPs and includes samples from 30 CEPH trios (CEU) from Utah, USA, with European ancestry; 30 Yoruban (YRI) trios from Ibadan, Nigeria; 45 unrelated Japanese subjects from Tokyo, Japan (JPT) and 45 unrelated Han Chinese from Beijing, China (CHB).
After filtering out SNPs with more than 5% missing data across samples and SNPs that were not in Hardy–Weinberg equilibrium in at least one of the analysis panels (P-value from exact test <0.001), and after removing three SNPs with allele coding errors, we obtained a subset of 102 208 SNPs that were typed in both the Saami and the HapMap samples. The median distance between adjacent SNPs was 10.1 kb and the first and third quartiles were 1.0 and 31.2 kb, respectively. Mean intermarker distance was 28.1 kb. After excluding SNPs that were monomorphic in at least one of the populations, 76 913 SNPs were shared between panels. The median intermarker distance for this map was 14.0 kb, with first and third quartiles 1.6 and 40.9 kb, respectively. Mean distance was 37.4 kb. NCBI build 36 coordinates were used throughout.
Evaluation of potential for genetic association studies
Estimation of genome-wide pairwise identity-by-descent (IBD) sharing, using a method of moments approach implemented in PLINK,21 revealed a substantial degree of undocumented relatedness among the Saami participants. Therefore, a subset of 100 maximally unrelated subjects was selected for the analysis with the aid of PedMine, which implements a simulated annealing algorithm24 (http://www.hg.med.umich.edu/labs/douglaslab/software.html). Within this subset, the maximum estimated genome-wide proportion of alleles shared IBD was 0.045, suggesting that subjects were not more closely related than second cousins. The sample size of 100 was chosen in order to have a sample that was roughly comparable in size to each of the HapMap reference panels.
We next compared minor allele frequency (MAF) distributions between Saami and HapMap panels. Due to their genetic similarity, the two Asian HapMap panels (JPT+CHB) were merged for all analyses. We calculated the correlation between allele frequency estimates for an arbitrarily chosen reference allele for 102 208 SNPs in the Saami sample and the CEU panel, and investigated whether there were instances of SNPs with MAF<5% in one sample and >10% in the other.
To evaluate the relative extent of LD we compared genome-wide average LD decay with genomic distance between the panels for common SNPs (MAF≥5% within panel). For each distance bin we calculated the proportions of SNP pairs with r2 and ∣D′∣≥0.80. In addition we used a sliding window approach: the averages of the LD measures r2 and ∣D′∣ were calculated for all SNPs within 500 kb from each other in sliding windows of 1.7 Mb (1.6-Mb overlap between adjacent windows). This choice of window size and SNP distance allows comparison with other studies.8 For this latter analysis we used the set of SNPs that were polymorphic in all panels. The LD statistics r2 and ∣D′∣ were calculated using Haploview25 (http://www.broad.mit.edu/mpg/haploview/) and further calculations were performed in R (http://www.r-project.org).
To compare long-range haplotype diversity in the Saami with the HapMap panels, we used the approach described by Service et al.8 In brief, the genome was divided into 1-Mb segments and segments containing ∼35 SNPs (range 30–40 SNPs) were retained for analysis. For each segment we inferred haplotypes and estimated their population frequencies using Haploview. Next we ranked haplotypes from common to rare, counted the number of haplotypes accounting for a given percentage of chromosomes and subsequently averaged this number over all segments. These calculations were performed in R.
Next, using PLINK we compared the abundance and length of extended regions of homozygosity (ROHs). The criteria used to identify such regions were length>1 Mb, at least 100 consecutive homozygous SNPs, with a density of minimum one SNP per 50 kb and no gap >1 Mb allowed. Also inbreeding coefficients were estimated as described by Purcell et al21 using a subset of SNPs in approximate linkage equilibrium (∼50 000 SNPs).
The relative power for genetic association studies in the Saami compared with that in the HapMap panels was quantified by determining the percentage of SNPs (of all ∼100 000) that had a proxy within 200 kb and within 2 Mb. The LD measure r2 was used to measure the correlation and its cut-off was varied along its entire range.
Results
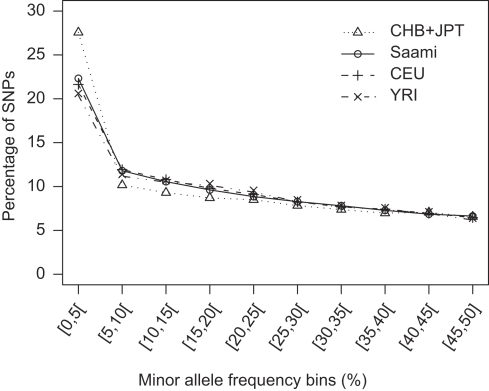
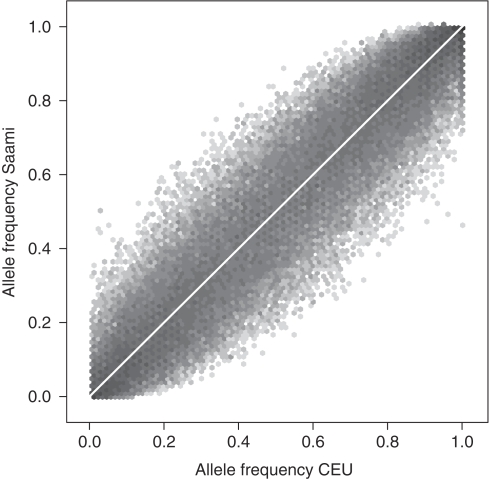
A comparison of the allele frequency distribution in the Saami with those in the HapMap panels indicates that the impact of the SNP ascertainment strategies is negligible (Figure 1). The MAF distribution in the Saami is very similar to that in the CEU panel, with a marginally higher proportion of SNPs with MAF<5%. The Pearson correlation between the allele frequency estimates in the Saami and those in the CEU panel was 0.97 (Figure 2). Using this set of 102 208 SNPs typed in both the Saami and HapMap panels, we also investigated whether there were instances of SNPs with MAF<5% in one sample and >10% in the other. Of 20 221 SNPs with MAF<5% in the CEU panel, we observed 1554 SNPs (7.7%) with frequency estimates ranging from 10.1 to 54.2% in the Saami. Conversely, out of 21 944 SNPs with MAF<5% in the Saami sample, 1453 SNPs (6.6%) with frequency estimates ranging from 10.2 to 31.7% were observed in the CEU panel.
Figure 1.
Allele frequency distributions. Comparison between the Saami and HapMap populations of minor allele frequency distributions for 102 208 SNPs.
Figure 2.
Comparison of allele frequency estimates in the Saami and the HapMap CEU panel. Allele frequency was estimated for an arbitrarily chosen reference allele for 102 208 SNPs. Only CEU founders were used. Hexagonal binning was used to visualize the results. Darker grey levels indicate that a greater number of points fall inside the hexagon.
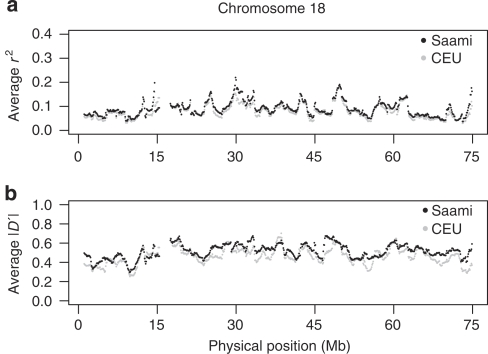
Next, using a sliding windows approach we compared the extent and patterns of LD along the entire genome between the different populations. Figure 3 shows an example comparison for chromosome 18. It can be seen that, on average, the LD measures r2 and ∣D′∣ are almost consistently higher in the Saami as compared with that in the CEU sample. This was most pronounced for ∣D′∣. This same pattern was observed for the other chromosomes (not shown). The distribution of LD for the CHB+JPT sample was very similar to that for the CEU sample (not shown).
Figure 3.
Comparison between the extent of LD on chromosome 18 in CEU and the Saami. The averages of the LD measures r2 (a) and ∣D′∣ (b) were calculated for all SNPs within 500 kb from each other in sliding windows of 1.7 Mb (1.6-Mb overlap between adjacent windows). Patterns of LD in the CHB+JPT panel were very similar to those in the CEU panel (not shown). Averages were almost consistently higher in the Saami as compared with that in the CEU and CHB+JPT panels. This was most pronounced for ∣D′∣. This same observation was made for all chromosomes (not shown).
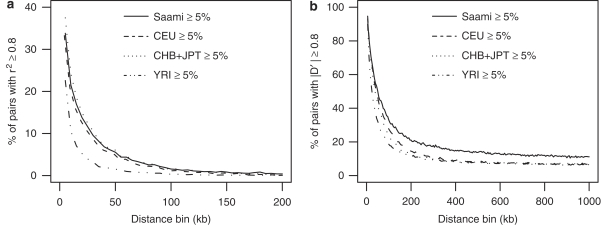
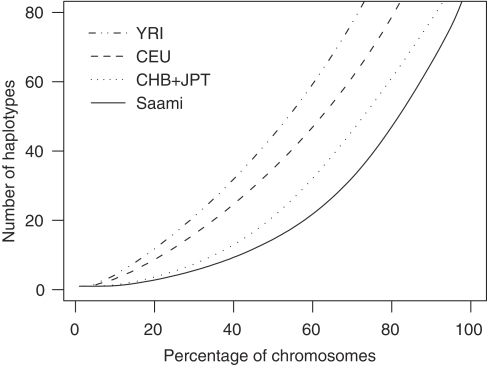
To investigate whether high LD is more frequent in the Saami, we compared the proportions of SNP pairs with r2 or ∣D′∣≥0.80 for a range of genomic distance bins for common SNPs (MAF≥5% within panel) between the different panels. For r2, the most relevant LD measure for genetic association studies because of its relationship with statistical power, the patterns of decay over distance are very similar for the Saami, CEU and CHB+JPT samples, the extent of high LD being marginally higher for distances <50 kb in CHB+JPT as compared with the CEU and Saami samples (Figure 4a). For statistic ∣D′∣, which measures historical recombination, the pattern was quite different. Here the proportion of marker pairs with high ∣D′∣ values was consistently higher in the Saami as compared to the other populations (Figure 4b). This observation can be ascribed to the reduced haplotype diversity as compared to the other populations. Figure 5 compares long-range haplotype diversity in the Saami with the HapMap panels. It can be seen that haplotype diversity is lowest in the Saami and that the diversity in CHB+JPT is lower as compared with that in CEU and YRI. The diversity is the highest in YRI. Among the Saami, a genome-wide average of 76 haplotypes accounted for 95% of the chromosomes, whereas 76 haplotypes accounted for 89% of the chromosomes among CHB+JPT, 79% among CEU and 69% among YRI. Considering the fact that the sample size for the Saami sample is larger than those for the HapMap panels (in particular the CEU and YRI samples, which are trios), the difference in haplotype diversity may even be more pronounced.
Figure 4.
Proportion of SNP pairs in high LD as a function of distance. For common SNPs (MAF≥5% within panel) the proportion of SNP pairs with values greater or equal to 0.80 was calculated for every distance bin using the LD statistics (a) r2 and (b) ∣D′∣.
Figure 5.
Haplotype diversity. The genome was divided into 1-Mb segments and segments containing ∼35 SNPs were retained for the analysis. Haplotypes were inferred and their population frequencies estimated for each segment. The number of haplotypes accounting for a given percentage of chromosomes was counted for every segment (starting counting from the most frequent haplotype). The average number of haplotypes over all segments is plotted against the percentage of chromosomes they accounted for.
The reduced haplotype diversity observed in the Saami is a consequence of their smaller historical population size. This is also reflected in a higher degree of background relatedness in the Saami as evidenced by comparison of the extent of homozygosity with the HapMap reference panels. We looked for extended ROHs in the genome and calculated the total length spanned by such regions. This analysis was performed on a subset of 100 maximally unrelated Saami subjects. Supplementary Figure 1 shows the results for the four analysis panels. Clearly, the Saami population is much more extreme in this respect, apart from three (known) outliers among the HapMap samples. For the Saami, median total ROH length was 61.3 Mb, with first and third quartiles 22.6 and 117.9 Mb, respectively. For CHB+JPT, CEU and YRI, median total length (first quartile; third quartile) was 5.0 Mb (2.5 and 8.6 Mb), 3.4 Mb (1.7 and 6.0 Mb) and 1.7 Mb (0 and 3.4 Mb), respectively. As expected, for the Saami, total ROH length was highly correlated with the inbreeding coefficient (Spearman rank correlation=0.89; Supplementary Figure 2). The median and maximum inbreeding coefficient for the Saami was 0.01 and 0.14, respectively. Note that the inbreeding coefficient estimates were calculated using an estimator based on genome-wide data. Hence, these values are not estimates of the classically defined inbreeding coefficient, which is derived from pedigree information.26
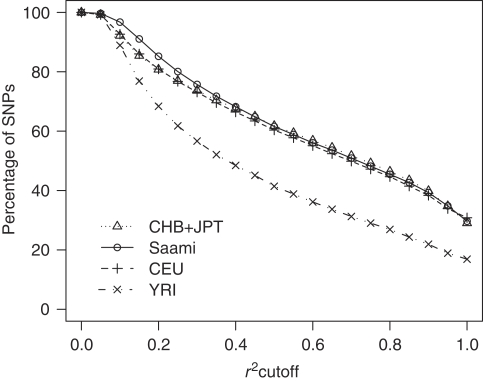
To quantify the relative power for genetic association studies in the Saami compared with that in the HapMap panels, we determined the percentage of SNPs (of all ∼100 000) that had a proxy within 200 kb and within 2 Mb. The LD statistic r2 was used to measure the correlation and its cut-off was varied along its entire range. The results for proxies within 2 Mb and within 200 kb were very similar, indicating that there is little long-range LD. Figure 6 (results for 2 Mb) shows that the difference in power between the Saami and the CHB+JPT and CEU panels is negligible. The percentage of SNPs that are highly correlated (r2≥0.80) to one or more others within 2 Mb for the CHB+JPT, CEU, Saami and YRI samples was 46.4, 44.7, 45.5 and 26.9%, respectively.
Figure 6.
Comparison of power for association studies. Relative power for genetic association studies in the Saami as compared with that in the HapMap panels was evaluated by determining the percentage of SNPs (of all ∼100 000) that had a proxy within 2 Mb, as measured by the LD statistic r2. The results for a 200-kb distance were very similar (not shown).
Discussion
This paper describes the results of an elaborate, genome-wide, SNP-based evaluation of the potential for GWASs for complex traits in the Finnish Saami. We studied the impact of SNP ascertainment strategies on the SNP frequency spectrum, extent and patterns of LD, haplotype diversity, and compared the power for association studies of common variants with that in the HapMap panels.
This study shows that patterns of LD in the Finnish Saami are very similar to those in the CEU and CHB+JPT HapMap reference panels. We found that, on average, the extent of LD is slightly higher in the Saami. Disappointingly, however, for an equivalent number of markers, genomic coverage as measured by the percentage of SNPs having a highly correlated proxy, does not differ much from that in the non-African HapMap panels. These results indicate that the potential for ‘drift mapping', anticipated based on simulations and limited empirical data, has been greatly overestimated.2, 9, 10, 13, 15, 16, 17
Most of the cited empirical work on the extent of LD in the Saami was based on microsatellite markers.2, 10, 15, 16 These studies only report P-values from pairwise significance tests of LD. Kaessmann et al13 also studied 50 SNPs in five genes, but only report D′-values. Pritchard and Przeworski27 clarified that r2 is the most pertinent measure for genetic association studies because of its direct connection with statistical power. In order to achieve roughly the same power at the marker locus as would be reached if the causative variant itself were tested, the sample size needs to be multiplied by 1/r2. The LD measures r2 and D′ behave very differently, and low values of r2 can be consistent with high values of D′. Indeed, our results demonstrate a relatively high extent of LD in the Saami, as measured by D′. However, this does not translate to elevated values for r2, that is, a power advantage. Our findings, thus, are not inconsistent with earlier studies reporting high values for D′.
Our findings, however, are in contrast with those of Johansson et al17 who reported dramatically elevated levels of LD as measured by r2 and a different basic LD structure. This latter study was based on array-based SNP discovery in a 4.4-Mb region of only 28 phased copies of chromosome 21 in the Swedish Saami. An explanation for this discrepancy could be that their region studied may not be representative for the rest of the genome. This is unlikely, however, based on the evidence that Johansson et al17 provide. Alternatively, genetic population substructure may have led to strong but artefactual LD. Recent SNP-based population genetic studies have exposed significant population substructure in Finnish early- and late-settlement subpopulations that correlates with geography.28, 29 Genetic substructure within the Saami is very likely given their linguistic and cultural diversity. Indeed, we found evidence for substructure within the Finnish Saami (results not shown). However, LD evaluation within subsets of our data defined by municipality, suggests that this did not appreciably affect LD estimates (results not shown). In general, genetic differentiation probably has to be extreme in order to affect LD estimates. The most plausible explanation is provided by Terwilliger and Weiss30 who show that a limited sample size can lead to upward biases of LD estimates. Johansson et al17 further state that there are serious limitations in the transferability of common tagSNPs between the HapMap and the Saami. The comparison they made, however, was unfair because the evaluation was not performed on the set of SNPs from which the tagSNPs were defined, a fact they acknowledge in their discussion. Based on their results, they suggest a difference in the basic correlation structure between Saami and the CEU HapMap population. Here, we show that this is not the case.
The pioneering simulation studies on the impact of demography on LD were based on multiallelic markers and only considered pairwise significance tests of LD.9, 10 Therefore, based on their results, little can be concluded about a power advantage for genetic association studies. Of course, real human populations differ from the idealized hypothetical populations upon which theoretical predictions are based. The precise demographic history of the Saami is uncertain. Pritchard and Przeworski27 already noted that the levels of genetic diversity at the microsatellite loci studied by Laan and Pääbo2 argue against the hypothesis of a very small population size. It could, thus, be that the effect of genetic drift on LD is minimal in the contemporary Saami population. In addition, in the absence of reference data for the Finns and other neighboring populations in Fennoscandia, admixture with those populations could not be investigated. This presents a limitation of our study.
In conclusion, our results indicate that the HapMap is a useful resource for genetic studies in the Finnish Saami. Imputation of untyped common SNPs, using a scaffold of LD relationships derived from the HapMap CEU panel, should be applicable in the Saami. We found that the power to detect common susceptibility alleles for common complex diseases in the Saami is similar to that in most other non-African populations. Thus, if the aim is to identify this class of variants, it seems that not much can be gained by conducting a genome-wide association scan or a candidate gene study in the Finnish Saami. This is especially true if one considers that several thousands of subjects need to be ascertained in order to reach sufficient power for detecting the typically small effect sizes for complex diseases.31 It seems unrealistic to recruit this large number of subjects in the Saami population. Furthermore, a higher extent of relatedness among subjects due to the smaller population size compromises the validity of classical statistical methods and requires more dedicated methodology32, 33 as compared with using an outbred population.
Recent studies demonstrate that complex disease susceptibility allele frequencies range from rare to common and that multiple rare variants within a single gene may contribute to common complex traits.34 Since the majority of rare SNPs are not represented on the early-generation SNP arrays, and since their study would require very large sample sizes, we can only draw very limited conclusions on the rare end of the SNP allele frequency spectrum. Our results indicate that some alleles that are rare in the CEU panel have drifted to frequencies in the Saami that would make them detectable in an association study. As usually, rare mutations arise on a different haplotypic background, the reduced haplotype diversity in the Saami may imply reduced allelic and genetic heterogeneity. Hence, in certain cases, the power for detecting associations with rare variants could well be higher in the Saami.
Acknowledgments
We express our most sincere gratitude to all the Saami volunteers who have participated in this study. This paper benefited from comments of two anonymous referees. This work was funded by the European Community (fifth Framework project QLRT-2001-00331), by the University of Antwerp (TOP project), by the Research Foundation – Flanders (FWO grant G.0163.09) and by the State of Arizona. JRH is a fellow of the Research Foundation – Flanders (FWO).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Peltonen L, Jalanko A, Varilo T. Molecular genetics of the Finnish disease heritage. Hum Mol Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- Laan M, Pääbo S. Demographic history and linkage disequilibrium in human populations. Nat Genet. 1997;17:435–438. doi: 10.1038/ng1297-435. [DOI] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M. Population choice in mapping genes for complex diseases. Nat Genet. 1999;23:397–404. doi: 10.1038/70501. [DOI] [PubMed] [Google Scholar]
- Eaves IA, Merriman TR, Barber RA, et al. The genetically isolated populations of Finland and Sardinia may not be a panacea for linkage disequilibrium mapping of common disease genes. Nat Genet. 2000;25:320–323. doi: 10.1038/77091. [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- Kristiansson K, Naukkarinen J, Peltonen L. Isolated populations and complex disease gene identification. Genome Biol. 2008;9:109. doi: 10.1186/gb-2008-9-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnen PE, Pe′er I, Plenge RM, et al. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat Genet. 2006;38:214–217. doi: 10.1038/ng1712. [DOI] [PubMed] [Google Scholar]
- Service S, DeYoung J, Karayiorgou M, et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Linkage disequilibrium in growing and stable populations. Genetics. 1994;137:331–336. doi: 10.1093/genetics/137.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Zöllner S, Laan M, Pääbo S. Mapping genes through the use of linkage disequilibrium generated by genetic drift: ‘drift mapping' in small populations with no demographic expansion. Hum Hered. 1998;48:138–154. doi: 10.1159/000022794. [DOI] [PubMed] [Google Scholar]
- Sajantila A, Lahermo P, Anttinen T, et al. Genes and languages in Europe: an analysis of mitochondrial lineages. Genome Res. 1995;5:42–52. doi: 10.1101/gr.5.1.42. [DOI] [PubMed] [Google Scholar]
- Lahermo P, Sajantila A, Sistonen P, et al. The genetic relationship between the Finns and the Finnish Saami (Lapps): analysis of nuclear DNA and mtDNA. Am J Hum Genet. 1996;58:1309–1322. [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Zöllner S, Gustafsson AC, et al. Extensive linkage disequilibrium in small human populations in Eurasia. Am J Hum Genet. 2002;70:673–685. doi: 10.1086/339258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajantila A, Pääbo S. Language replacement in Scandinavia. Nat Genet. 1995;11:359–360. doi: 10.1038/ng1295-359. [DOI] [PubMed] [Google Scholar]
- Laan M, Pääbo S. Mapping genes by drift-generated linkage disequilibrium. Am J Hum Genet. 1998;63:654–656. doi: 10.1086/301972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A, Vavruch-Nilsson V, Edin-Liljegren A, Sjolander P, Gyllensten U. Linkage disequilibrium between microsatellite markers in the Swedish Sami relative to a worldwide selection of populations. Hum Genet. 2005;116:105–113. doi: 10.1007/s00439-004-1213-8. [DOI] [PubMed] [Google Scholar]
- Johansson A, Vavruch-Nilsson V, Cox DR, Frazer KA, Gyllensten U. Evaluation of the SNP tagging approach in an independent population sample – array-based SNP discovery in Sami. Hum Genet. 2007;122:141–150. doi: 10.1007/s00439-007-0379-2. [DOI] [PubMed] [Google Scholar]
- Fransen E, Topsakal V, Hendrickx JJ, et al. Occupational noise, smoking, and a high body mass index are risk factors for age-related hearing impairment and moderate alcohol consumption is protective: a European population-based multicenter study. J Assoc Res Otolaryngol. 2008;9:264–276. doi: 10.1007/s10162-008-0123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laer L, Van Eyken E, Fransen E, et al. The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum Mol Genet. 2008;17:159–169. doi: 10.1093/hmg/ddm292. [DOI] [PubMed] [Google Scholar]
- Huyghe JR, Van Laer L, Hendrickx JJ, et al. Genome-wide SNP-based linkage scan identifies a locus on 8q24 for an age-related hearing impairment trait. Am J Hum Genet. 2008;83:401–407. doi: 10.1016/j.ajhg.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JA, Sandefur CI. PedMine – a simulated annealing algorithm to identify maximally unrelated individuals in population isolates. Bioinformatics. 2008;24:1106–1108. doi: 10.1093/bioinformatics/btn087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Malécot G. Les Mathématiques de l′Hérédité. Paris: Masson; 1948. [Google Scholar]
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakkula E, Rehnstrom K, Varilo T, et al. The genome-wide patterns of variation expose significant substructure in a founder population. Am J Hum Genet. 2008;83:787–794. doi: 10.1016/j.ajhg.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela E, Lappalainen T, Fransson I, et al. Genome-wide analysis of single nucleotide polymorphisms uncovers population structure in Northern Europe. PLoS ONE. 2008;3:e3519. doi: 10.1371/journal.pone.0003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Weiss KM. Linkage disequilibrium mapping of complex disease: fantasy or reality. Curr Opin Biotechnol. 1998;9:578–594. doi: 10.1016/s0958-1669(98)80135-3. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Amin N, van Duijn CM, Aulchenko YS. A genomic background based method for association analysis in related individuals. PLoS ONE. 2007;2:e1274. doi: 10.1371/journal.pone.0001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.