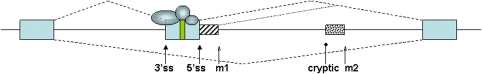
Figure 1.
Schematic overview over disease mechanisms. A hypothetical pre-mRNA is shown. Exons are indicated as boxes, introns as lines. The splicing pattern is depicted by dashed lines. Proteins that aid in exon recognition are indicated as circles. Only three proteins are shown for simplicity. Most of the pre-mRNA will be coated with proteins. An exonic regulatory element is shown by a green rectangle in the alternative exon. Mutations in this exon can alter the usage of the alternative exon and lead to a human disease, even if the mutation does not change the predicted reading frame. The 3′ and 5′ splice sites of the alternative exon (3′ss and 5′ss, respectively) are shown by solid pointed arrows. Mutations of these splice sites generally change exon usage and in most cases studied abolish the use of the alternative exon. In addition, intronic mutations that are indicated by open arrows can generate new exons, if these mutations generate new splice sites. An intronic mutation m1 that is close to the authentic 5′ splice site can lead to the generation of a cryptic exon that is indicated by a striped, smaller box and its splicing pattern is indicated by a dotted line. The deep intronic mutation m2 can lead to the formation of a new exon if it is close enough to a cryptic splice site, indicated by a round arrow. For simplicity, only mutations leading to new 5′ splice sites are shown, but new 3′ splice sites can also be generated by new mutations.