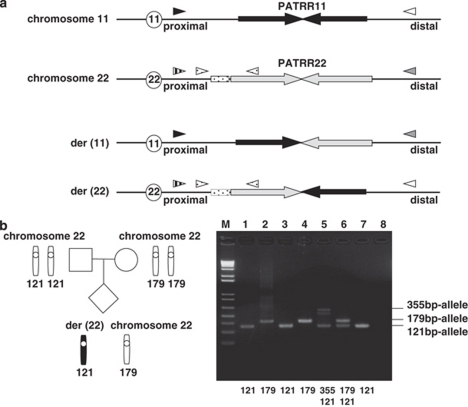
Figure 1.
The PCR system to determine the allele type of the translocated or normal chromosomes. (a) Translocation-specific PCR. Black arrows indicate each proximal and distal unit of the PATRR11, whereas light gray arrows depict the PATRR22. The PCR primers are indicated as arrowheads. The PATRRs on the normal chromosomes were amplified with a PATRR11-specific primer set (black and white arrowheads) or that for PATRR22 (vertical-striped and gray arrowheads). Translocations can be detected using one primer flanking the PATRR11 (black or white arrowheads) and one primer flanking the PATRR22 (vertical-striped or gray arrowheads). Stippled boxes indicate the AT-rich regions flanking the PATRR22. To determine the size of the AT-rich region, we performed nested PCR. The first PCR amplified the PATRR22 or der(22) breakpoint region using specific-primer sets, respectively. Next, internal primers (stippled arrowheads) were used to amplify the AT-rich regions flanking the PATRR22. (b) Determination of the parental origin in family 6. PCR was performed to amplify the AT-rich region adjacent to the PATRR22. The figure shows the ideograms for the normal chromosome 22, and the der(22). M: 1 kb Plus size marker, lane 1: the der(22) of the translocation carrier, lane 2: chromosome 22 of the carrier, lane 3: father, lane 4: mother, lane 5: heterozygote for the 355 bp- and 121 bp-alleles 6: heterozygote for the 179 bp- and 121 bp-alleles, 7: homozygote for the 121 bp-alleles, 8: no DNA. The additional band observed in lane 5 originates from the heteroduplex. Allele type of the der(22) of the carrier corresponds to the paternal type, and that of the PATRR22 of the normal homolog to the maternal type.