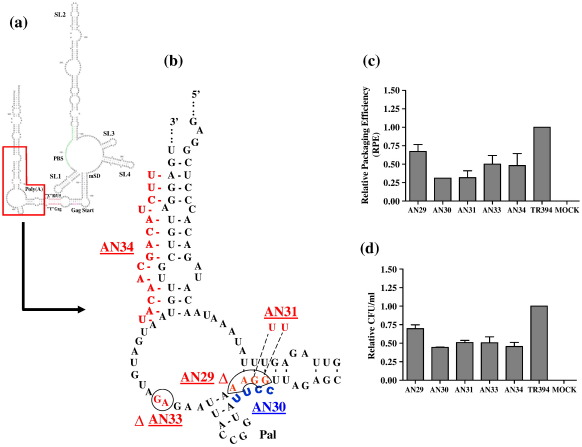
Fig. 6.
A representation of the deletion/substitution mutations introduced in the gag sequences base pairing with the 5′ end of the RNA genome in the predicted RNA secondary structure and their effect on transfer vector RNA packaging and propagation. (a) FIV RNA secondary structure highlighting the region showing the base pairing between the 5′ and 3′ ends of FIV RNA packaging determinants. (b) Enlarged view of the boxed area in Fig. 5a showing the specific mutations that were introduced. (c) RPEs of the transfer vectors containing deletion/substitution mutations in gag. (d) Relative transfer vector RNA propagation for each mutant expressed as Hygr CFU/ml of the viral supernatant and the data were derived after normalization to the transfection efficiency. The data are derived from at least three independent transfection and infection experiments except for the RPEs of AN30. Poly(A), polyadenylation sequence; PBS, primer-binding site; mSD, major splice donor; Pal, palindromic sequence.