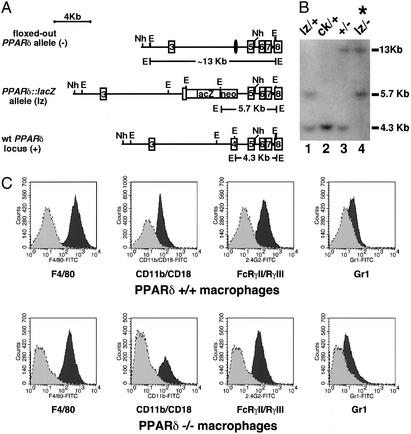
Figure 2.
Generation of PPARδ-deficient ES cells and macrophages. (A) Targeting scheme for generation of PPARδ null ES cells. The G418-sensitive ES cell clone IIIA4 (23) consists of a mixture of cells heterozygous for the WT PPARδ allele (+, Bottom) and either a null “floxed-out” allele (−, Top) or a conditional “floxed” allele (ck, not shown). This clone was retargeted with a construct containing a lacZ gene knocked-in, which disrupts PPARδ upstream of the first zinc finger DNA-binding motif (lz, Middle). Patterned bars below each allele represent the corresponding predictions of EcoRI-flanked DNA fragments when probed with a 3′-external probe. Restriction sites: E, EcoRI; Nh, NheI. (B) Southern blot analysis of PPARδ null ES cells. The individual daughter clones generated by retargeting the IIIA4 cells contain different biallelic combinations of PPARδ: cells heterozygous for the WT and the newly introduced lacZ knock-in allele (+/lz; lane 1); two G418-resistant clones carrying the original IIIA4 allelic combinations ck/+ and +/− (lanes 2 and 3, respectively) and a likely nonhomologous integration of the neoR gene; and the PPARδ null ES clone analyzed functionally in this study (lz/−, lane 4; *), in which the lz allele integrated in place of the remaining WT allele of clone IIIA4. Allelic identity was further confirmed using a 5′ external probe (not shown). (C) FACS analysis of PPARδ+/+ and PPARδ−/− ES cell-derived macrophages. Macrophages were generated from ES cells as described in Materials and Methods. WT or PPARδ null macrophages were stained with the indicated antibodies (dark gray) or isotype control (light gray). Macrophage cell-surface markers include F4/80, Mac-1 (CD11b/CD18), and 2.4G2 (FcγRII/FcγRIII). Gr.1 (Ly-6G), a granulocyte lineage-specific marker, is used here as a negative control.