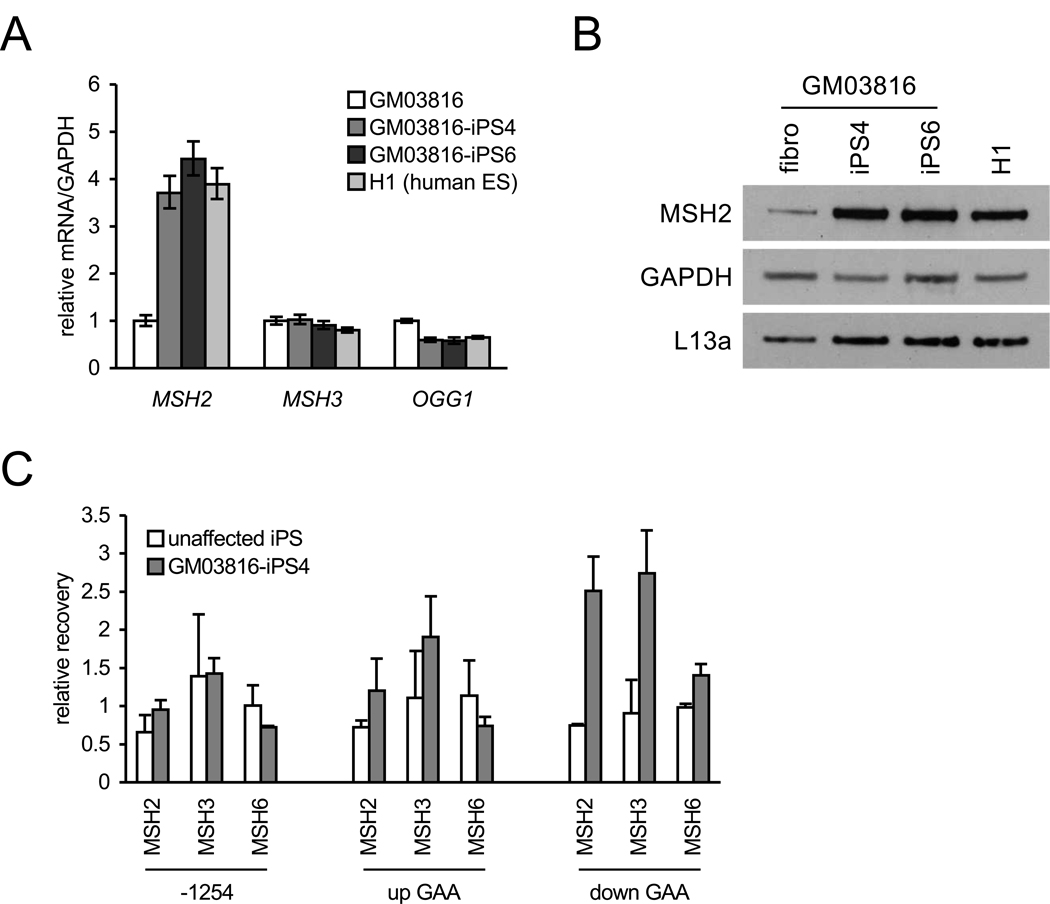
Figure 3. MSH2 expression and localization in FRDA iPSCs.
(A) MSH2, MSH3, and OGG1 mRNA levels in GM03816 fibroblasts (white), two GM03816 iPSC lines (iPS4, medium grey; iPS6, dark grey) and H1 cells (light grey). Levels are normalized to GAPDH. Errors bars = SEM of duplicate measurements.
(B) Western blot of MSH2 protein. Left to right: GM03816 fibroblast, two GM03816 iPSC lines (4 and 6), and H1 hESCs. GAPDH and L13a are loading controls.
(C) MSH2, MSH3, and MSH6 occupancy on the FXN gene 1254 bp upstream of the transcriptional start site (−1254), upstream (up GAA) and downstream (down GAA) of the GAA•TTC repeat expansion (measured by ChIP). IP recovery is relative to FXN intron 2 (11482 bp downstream of the transcriptional start site). Error bars = SEM of independent experiments (MSH2, n = 4; MSH3 and MSH6, n = 2).