Abstract
Vibrio cholerae is the etiologic bacterial agent of cholera, a severe diarrheal disease endemic in much of the developing world. The V. cholerae genome contains 3,890 genes distributed between a large and a small chromosome. Although the large chromosome encodes the majority of recognizable gene products and virulence determinants, the small chromosome carries a disproportionate number of hypothetical genes. Thus, little is known about the role of the small chromosome in the biology of this organism or other Vibrio species. We have used the rabbit ileal loop model of V. cholerae infection to obtain in vivo-grown cells under near midexponential conditions in the small-intestinal environment. We compared the global transcriptional pattern of these in vivo-grown cells to those grown to midexponential phase in rich medium under aerobic conditions. Under both conditions, the genes showing the highest levels of expression reside primarily on the large chromosome. However, a shift occurs in vivo that results in many more small chromosomal genes being expressed during growth in the intestine. Our analysis further suggests that nutrient limitation (particularly iron) and anaerobiosis are major stresses experienced by V. cholerae during growth in the rabbit upper intestine. Finally, relative to in vitro growth, the intestinal environment significantly enhanced expression of several virulence genes, including those involved in phenotypes such as motility, chemotaxis, intestinal colonization, and toxin production.
The environmental bacterium Vibrio cholerae is the causative agent of cholera, a severe diarrheal disease endemic in much of South Asia, Africa, and Latin America (1). Clones of V. cholerae that emerge to cause epidemic and pandemic disease do so through acquisition of accessory genetic elements such as phages and chromosomal pathogenicity islands that encode key virulence factors such as cholera toxin (CT) (2) and the intestinal colonization factor, toxin-coregulated pilus (TCP) (3). The expression of these two coordinately regulated virulence factors has been extensively studied both in vitro and in vivo (4, 5). However, we know little about the metabolic pathways that are critical to the replication of V. cholerae during infection (6, 7).
The genome of V. cholerae strain N16961 consists of two circular chromosomes, one large (2.96 × 106 bp) and one small (1.07 × 106 bp), which respectively encode 2,775 and 1,115 ORFs (8). The large chromosome encodes the majority of recognizable “housekeeping” gene products involved in transcription, translation, metabolism, and cell biology, whereas the small chromosome encodes many more hypothetical gene products. Recently, the V. cholerae genome was scanned genetically for genes encoding “essential gene products” defined as those required for optimal growth on rich laboratory media (9). This analysis revealed that the majority of essential genes reside on the large chromosome. Thus, the biological role of the small chromosome remains largely speculative.
We recently constructed a genomic V. cholerae microarray that we used in comparative genomics (10) and to expression profile regulatory mutants affected in quorum sensing and virulence (11). More recently, Merrell et al. (12) reported microarray transcriptional profiling of bacteria shed in the stools of cholera patients. Although cholera patients are an interesting source of “in vivo”-grown V. cholerae, they may not be the ideal source for experimental purposes, because it is difficult to control the time points for harvesting bacteria from cholera patients, who are a diverse group (varying in age, sex, nutritional and immune status, diet, etc.), and who arrive at clinics in different stages of the illness. Also, bacteria recovered from stools may be in a different physiological state(s) than bacteria growing in the upper intestine (where replication and pathogenesis occurs) because of their transit through the large intestine.
To avoid these limitations, we used the rabbit ileal loop model of V. cholerae infection to obtain in vivo-grown cells under near midexponential conditions in the small-intestinal environment. Here we report the global transcriptional pattern of these in vivo-grown cells compared with those grown under laboratory conditions.
Materials and Methods
Strains and Growth Conditions.
V. cholerae strain N16961 (El Tor, O1, StrR) was used for this study. For midexponential phase RNA preparation, this strain was grown at 37°C in 50 ml of LB on a rotary shaker at 250 rpm.
Rabbit Ileal Loop Model and Isolation of in Vivo-Grown Bacteria.
The ligated rabbit intestinal loop model was performed essentially as described (13) by using New Zealand rabbits (female, ≈6 mos). V. cholerae cells (105, grown aerobically in LB) suspended in PBS with 1% BSA were inoculated into each 10-cm ligated segment. Two loops per rabbit were injected with saline as negative controls. After 8 h, the rabbits were killed, and the small intestine was removed. Fluid within the loops was collected separately into tubes on ice and the viable counts determined by plating on LB agar. Bacteria were harvested from ice-cold loop fluid by centrifugation and used for preparation of RNA within 1 h of collection.
DNA and RNA Preparation, Labeling, and Hybridization.
Genomic DNA and RNA were prepared from V. cholerae cells in mid-log phase (OD600 0.3–0.4) by using the Easy-DNA kit (Invitrogen) and TRIzol reagent (GIBCO/Life Technologies), respectively. Total RNA was treated with DNase I (Ambion, Austin, TX). Genomic DNA (gDNA) was labeled with Cy5 or Cy3-dCTP (Perkin–Elmer Life Sciences), as described (10). RNA was reverse-transcribed to produce fluorescently labeled cDNA (11). After purification using the Qiagen (Chatsworth, CA) PCR purification kit, the labeled cDNA probes were combined with labeled gDNA probes and applied to the V. cholerae microarray for the “RNA vs. gDNA” experiments. For “RNA vs. RNA” experiments, differentially labeled cDNA probes derived from RNA from in vivo- and in vitro-grown cells were mixed and hybridized to the arrays. The V. cholerae microarray has been described and consists of full-length PCR products of the 3,890 genes (10). The procedures for probe hybridization, washing of arrays, and data collection have been described (10).
Data Analysis.
For each growth condition analyzed, data were derived from two to four microarray experiments. Two independent RNA preparations were used for the in vitro experiments, and three from vibrios harvested from each of three different rabbit ileal loops were used for the in vivo study. Chromosomal DNA probe was used as an internal reference in each hybridization experiment. As discussed elsewhere (14, 15), we expected that this “RNA vs. gDNA” method would help normalize differences in the loading of PCR products on the microarray or hybridization kinetics due to variability in the size or GC content of any given gene. To avoid variation of Cy3 and Cy5 incorporation, both Cy3- and Cy5-labeled cDNA and genomic DNA were used for hybridizations. Ratios of hybridization signals of cDNA vs. genomic DNA probes on each spot were used to represent the expression level of each gene.
Fluorescence intensity data from each array were collected with a ScanArray 5000 scanner (Packard) and GENEPIX PRO 4.0 software (Axon, Foster City, CA). During scanning, the Cy5 and Cy3 channels were balanced by adjusting laser power and the photo multiplier tube. Initial array analysis included only those spots meeting standards previously described (10, 11), and calculations were performed by using background-corrected fluorescence intensity values. The genespring software package from Silicon Genetics (Redwood City, CA) was used for further analysis of the microarray data.
RT-PCR.
RNA isolated from bacteria from ileal loops was treated with DNase I and then reverse-transcribed to cDNA by using random hexamers, as described above. PCR was then performed by using cDNA templates and primers specific for V. cholerae genes VC2187, VC2033, VCA0933, VC0844, VC0769, and VCA0853 (8). Control PCR reactions were performed by using chromosomal DNA template and the same primer sets under the same PCR conditions.
Results
The Transcriptome of V. cholerae Growing Aerobically in Vitro.
To determine gene expression profiles of V. cholerae cells under in vitro conditions, we used an “RNA vs. gDNA” approach with our V. cholerae microarray (10) to measure mRNA levels of midexponential phase N16961 cells grown aerobically in LB. In brief, cDNA derived from RNA prepared from cells grown under these conditions was hybridized to microarrays in the presence of a differentially labeled reference gDNA probe derived from chromosomal DNA prepared from midexponential phase cells.
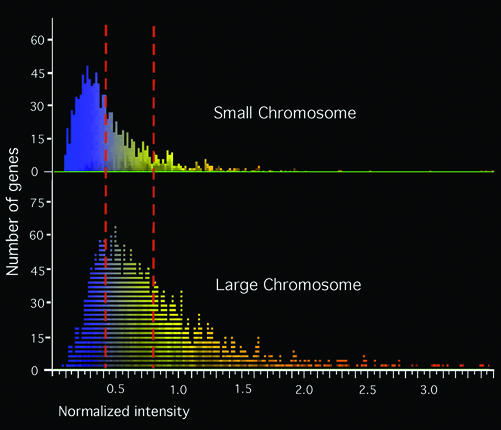
The expression levels of 3,890 genes were analyzed (Data Set 1, which is published as supporting information on the PNAS web site, www.pnas.org) and the results graphically displayed relative to their chromosomal location (Fig. 1). The genes showing the highest expression levels tended to be located on the large chromosome, whereas the small chromosome carried a disproportionate number of the genes showing the lowest expression levels. Genes were rank ordered from those showing the highest to those showing the lowest expression (Table 1). Of the 3,890 V. cholerae genes, 1,115 are located on the small chromosome. If gene expression levels were randomly distributed, the expected number of small-chromosome genes in any arbitrarily selected range would be 1,115/3,890 or 28.7%. However, the actual number of small-chromosome genes in any given range of expression level was lower than expected, except for genes showing the lowest levels of expression (Table 1). For example, of the one-third of genes showing the lowest expression (the range from 2,594 to 3,890), 50.6% are located on the small chromosome. Thus, the small chromosome not only lacks the genes that show the highest levels of expression in LB, but it is also disproportionately enriched in genes showing the lowest levels of expression.
Figure 1.
Graphical representation of V. cholerae gene expression in LB. (Upper) Distribution of small-chromosome genes. (Lower) Distribution of large-chromosome genes. Three thousand eight hundred-ninety genes were analyzed by using GENESPRING, and the expression levels of these genes are represented by normalized intensities. On the basis of expression levels, dashed lines divide the total genes into three equal areas: top one-third, middle one-third, and bottom one-third. One hundred seven genes, whose expression levels were >3.5, are not listed.
Table 1.
Comparison of gene expression levels between small and large chromosomes of V. cholerae
| Range of genes* | Percentage (number) of small chromosomal genes
|
Percentage (number) of large chromosomal genes
|
||
|---|---|---|---|---|
| LB | Ileal loop | LB | Ileal loop | |
| 1–100 | 3.0 (3) | 16.0 (16) | 97.0 (97) | 84.0 (84) |
| 101–200 | 6.0 (6) | 18.0 (18) | 94.0 (94) | 82.0 (82) |
| 201–300 | 6.0 (6) | 23.0 (23) | 94.0 (94) | 77.0 (77) |
| 301–600 | 14.0 (42) | 18.3 (55) | 86.0 (258) | 81.7 (245) |
| 601–900 | 14.3 (43) | 19.3 (58) | 85.7 (257) | 80.7 (242) |
| 901–1,296 | 15.4 (61) | 22.0 (87) | 84.6 (335) | 78.0 (309) |
| 1,297–2,593 | 22.9 (297) | 27.1 (351) | 77.1 (1,000) | 72.9 (946) |
| 2,594–3,890 | 50.6 (657) | 39.1 (507) | 49.4 (640) | 60.9 (790) |
Genes are listed in the order of their gene expression levels, and the gene with the highest expression level is on the top of the list.
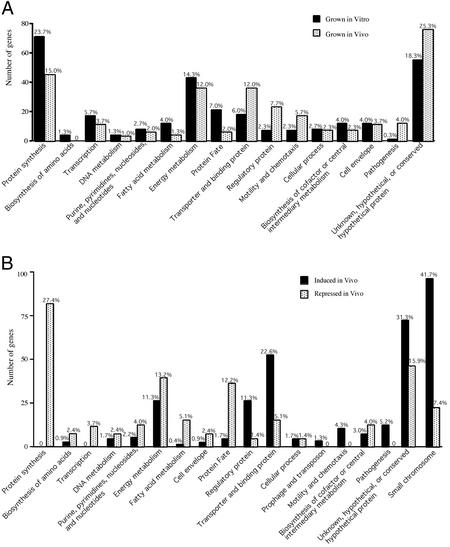
We selected the 300 V. cholerae genes showing the highest expression levels in LB (see Table 2, which is published as supporting information on the PNAS web site) and categorized these by chromosomal location and function (Fig. 2A). As observed for the entire in vitro transcriptome (Fig. 1), 285 of the 300 most highly expressed genes resided on the large chromosome. The largest functional group contains 71 genes (23.7% of the 300) involved in protein synthesis and accounted for 55.0% of 129 known genes in the translation apparatus (Fig. 2A). These 71 genes included 54 of the 59 known genes encoding ribosomal proteins, and the remaining 17 genes encoded tRNA synthetases and translation factors. These results are consistent with the fact that rapidly growing cells require high levels of protein synthesis (16).
Figure 2.
Functional classes of differentially expressed V. cholerae genes. (A) Functional categories of the 300 genes with the highest expression levels in vitro (aerobic growth in LB) and in vivo (growth in rabbit ileal loops). (B) Functional categories of genes showing 2-fold or greater changes under in vivo compared with in vitro growth conditions. The percentage of genes in each category appears above each bar.
Only one gene in the pathogenesis category was among the 300 genes most highly expressed in LB. This gene (VC1130) encodes the DNA-binding protein VicH, which regulates motility (17). Other genes involved in production of TCP or CT were not highly expressed. Thus, growth in midexponential phase in LB is not optimal for virulence gene expression for N16961, consistent with previous observations (18). It is worth noting, however, that the ToxR-regulated gene ompU was among the five most highly expressed genes (see Data Set 1), consistent with previous observations that OmpU is apparently the most abundant protein expressed by V. cholerae grown in LB (19).
Somewhat surprising was the fact that many highly expressed genes were in the group annotated as unknown, conserved hypothetical, and hypothetical genes (Fig. 2A). This was the second largest group, representing 18.3% (55 genes) of the 300 most highly expressed genes. Six of these 55 genes are duplicated or present in multiple copies (VC0314, VC0713, VC0160, VC0388, VC2495, and VC2752). Another gene (VC2155) was present in single copy and was also the most highly expressed hypothetical gene. These seven genes are relatively small, varying in size between 93 and 195 bp, and are located immediately next to ribosomal RNA genes. Thus, readthrough transcription from rRNA genes may account for the high-level expression observed for these particular small hypothetical ORFs.
Preparation of in Vivo-Grown Cells and Measurement of in Vivo Expression.
We selected the rabbit ileal loop model to obtain in vivo-grown V. cholerae for transcriptome analysis. Because we hoped to compare exponential-phase in vivo-grown bacteria to exponential-phase in vitro-grown bacteria, we performed experiments to determine how to harvest exponential-phase cells from loops. Empirically, we determined that inoculating between 104 and 105 colony-forming units (cfu) of strain N16961 into an ileal loop and then harvesting bacteria from the loop 8 h later provided a yield of nearly 108 cfu per milliliter of loop fluid. Because other loops inoculated with 10 times more organisms yielded between 109 and 1010 cfu per milliliter at 8 h, we concluded that cells harvested from 8-h loops, which yielded 108 cfu per milliliter represented midexponential phase cells grown in vivo. Transcriptional profiling analysis of cells harvested from loops yielding higher levels of organisms supports this conclusion (see Discussion).
As a control, we also inoculated ileal loops with only PBS. No bacteria were detected in fluid harvested from these loops. Nonetheless, we prepared RNA from fluid from these control loops to detect any source of background signal arising from rabbit nucleic acids or nonculturable normal bacterial flora. We detected no hybridization signal when our V. cholerae genomic microarray was hybridized to probe derived from control loop RNA preparations.
To analyze the transcriptional state of in vivo-grown V. cholerae harvested from rabbit ileal loops, we used two complementary approaches. First, we used the “RNA vs. gDNA” method described above for analysis of in vitro-grown bacteria. We also performed “RNA vs. RNA” cohybridization experiments, in which differentially labeled cDNA probes derived from RNA from in vivo- and in vitro-grown cells were mixed and hybridized to the arrays. The trends and specific examples discussed below were consistently observed with both experimental approaches (see Data Sets 2 and 3, which are published as supporting information on the PNAS web site).
The Transcriptome of V. cholerae in Vivo.
The overall in vivo gene expression pattern of V. cholerae is summarized in Table 1 as a rank ordering of all genes by absolute expression level, which was determined by using genespring analysis of RNA vs. gDNA data. In the top one-third of all expressed genes (1–1,296), the ratio of small-chromosome genes remains 16.0–23.0%, lower than the expected random ratio of 28.7%. For the middle one-third (1,297–2,593), the ratio increases to 27.1%, close to the random ratio, and in the bottom one-third (2,594–3,890), the ratio further increases to 39.1%. Overall, these results indicate that, as for in vitro-grown cells, large-chromosome genes were generally expressed at higher levels in vivo than small-chromosome genes. However, there was a strong trend toward expression of many more small-chromosome genes for in vivo-grown cells compared with in vitro-grown cells. For example, the differences between the loops and LB are most obvious in the ranges 1–100, 101–200, and 201–300. Highly expressed small-chromosome genes represented only 3–6% for LB-grown cells but 16–23% for cells from ileal loops.
We classified the 300 genes that were most highly expressed in vivo (see Table 3, which is published as supporting information on the PNAS web site) by functional group (Fig. 2A). Again, the genes most highly expressed in vivo resided on the large chromosome, but many more small-chromosome genes were scored as highly expressed. The general distribution of genes among the various functional groups was similar to that obtained for in vitro-grown cells, suggesting that we successfully harvested in vivo-grown bacteria in midexponential phase. However, there was a trend that fewer genes showing the highest levels of expression in LB also showed the highest levels of expression in vivo. This trend was apparent in most “housekeeping” categories such as protein synthesis (45 in vivo vs. 71 in vitro), transcription (11 vs. 17), fatty acid metabolism (4 vs. 12), and protein fate (6 vs. 21). These trends could reflect either a slower growth rate in vivo compared with in vitro, an increase in expression of genes poorly expressed in LB, a mixed population of cells in various stages of growth, or a combination of these effects.
In fact, a substantial metabolic shift is indicated by the genes expressed at the highest levels in vivo (Fig. 2A and supporting information on the PNAS web site). For example, 36 energy metabolism genes (12.0% of the 300) were highly expressed in vivo, but only 17 of these were among the top 300 genes expressed in LB. Of these 36 in vivo-expressed genes, 24 are involved in carbohydrate metabolism, and 11 encode enzymes involved in anaerobic energy metabolism, including fumarate reductases (frdABCD), alcohol dehydrogenase (adhE), pyruvate formate lyase (pflB), and glycerol-3-dehydrogenases (glpBC). Five of these genes involved in anaerobic metabolism were among the 30 genes most highly expressed in vivo. Of the remaining energy-related genes, seven encode electron transporters, including cytochrome c-type protein (yecK). These results suggest that anaerobic conditions exist in the ileal loop environment and that V. cholerae may derive much of its energy from anaerobic respiration using alternative electron acceptors such as fumarate, or donors such as formate or glycerol-3-phosphate. We also found three genes involved in amino acid catabolism, and one of these, aspA, is among the 30 genes most highly expressed in vivo. aspA encodes aspartase, which converts aspartate to fumarate and ammonium ion, suggesting that aspartate may serve as a source of nitrogen as well as fumarate for use as an alternative anaerobic electron acceptor within the gut environment.
Another functional group showing a large increase in expression during in vivo growth was transporters and binding proteins, which contained 36 (12.0%) of the 300 most highly expressed genes (Fig. 2A). These included genes encoding transporters of carbohydrates, organic acids, cations, peptides, amino acids, purines, and pyrimidines. The three anaerobic C4-dicarboxylate transporter genes (dcuABC) were among these and are consistent with the anaerobic scavenging of fumarate (20, 21). There was also a clear trend suggesting that genes involved in iron acquisition were highly expressed in vivo. These included genes involved in the synthesis, binding, and transport of vibriobactin, the major iron (III) siderophore of V. cholerae (22). Two genes involved in iron (II) transport (feoAB) were also highly expressed, as were genes encoding an iron (III) and hemin ABC transporter complexes. These results indicate that V. cholerae cells are starved for iron within the rabbit upper intestine, a conclusion that was also reached in a previous study of in vivo-expressed outer membrane proteins (23).
Several other functional categories showed substantial increases in relative expression compared with in vitro-grown cultures, including regulatory proteins, motility/chemotaxis, and pathogenesis (Fig. 2A). Of the 300 genes most highly expressed in vivo, 12 (compared with only one in LB) belonged to the pathogenesis functional group. These included the virulence regulators tcpP, tcpH, and toxR (24, 25). The hemagglutinin protease gene (hap) was highly expressed despite the fact that the regulatory gene (hapR) that controls its expresssion is mutated in N16961 (11). In addition, genes for the hemolysin HlyA and its transporter HlyB, the mannose-sensitive hemagglutinin (MSHA) type IV pilus (mshABCD), and many motility/chemotaxis genes were highly expressed. Both the MSHA pilus adhesin and motility greatly stimulate adherence to both inert surfaces and in vivo ligands such as intestinal mucus. Of the 11 genes in the cell envelope group, five encoded outer membrane proteins (including the ToxR-regulated ompu gene) or lipoproteins. Like LB-grown cells, in vivo-grown cells expressed many lipopolysaccharide and O antigen biosynthesis genes at very high levels.
Finally, 25.3% (76) of the 300 genes highly expressed encode unknown, hypothetical and conserved hypothetical proteins (Fig. 2A). Six of the seven hypothetical genes most highly expressed in LB (VC0314, VC0713, VC2155, VC0388, VC2495, and VC2752) are also highly expressed in vivo. As noted above, transcriptional activation of the small chromosome in vivo is apparent even among the most highly expressed genes (Fig. 2A), and many of these genes encode unknown, hypothetical, or conserved hypothetical proteins.
Validation of Selected in Vivo Expression Results.
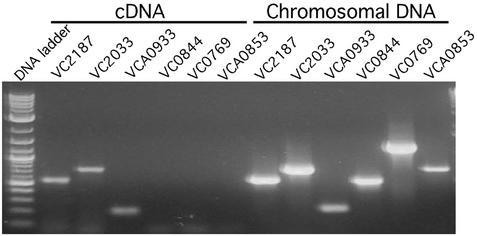
To verify the in vivo microarray data, we performed RT-PCR on RNA prepared from in vivo-grown bacteria, using primers specific for selected V. cholerae genes (VC2187, VC2033, VCA0933, VC0844, VC0769, and VCA0853, encoding FlaC, alcohol dehydrogenase, cold-shock domain family protein, AcfA, a putative chitinase, and a hypothetical protein, respectively). Our array data indicated that the first three of these genes were highly expressed, whereas others were poorly or not expressed. RT-PCR specific for the three highly expressed genes gave products of the same size as those generated from a positive control experiment (Fig. 3). In contrast, RT-PCR specific for the three poorly expressed genes produced no product from RNA from in vivo-grown cells. To ensure that the absence of PCR product was not due to problems with the primers, different sets of primers were used for these reactions. No PCR products were obtained with any primer set. When chromosomal DNA was used as a control template, all three poorly expressed genes gave products of the expected size, indicating that the PCR conditions were robust and that there was no DNA contamination in our RNA samples. The consistency of the RT-PCR analysis with the microarray results suggests that our conclusions regarding expression levels of various genes are accurate.
Figure 3.
RT-PCR of total RNA isolated from rabbit ileal loops. PCR was performed as described in Materials and Methods by using primers specific for selected V. cholerae genes and either reverse-transcribed RNA (cDNA) or chromosomal DNA as template.
Comparison of Gene Expression Between LB and Rabbit Ileal Loops.
We wished to determine which genes showed the most dramatic changes in expression in the ileal loop compared with LB. This was done by examining trends apparent in multiple data sets generated by two different experimental approaches as described above (i.e., RNA vs. gDNA, and RNA vs. RNA). Using genespring analysis of the RNA vs. gDNA, and RNA vs. RNA data, we found 230 genes with significantly higher expression (at least 2-fold) in cells derived from ileal loops compared with in vitro growth, and 296 genes with at least a 2-fold decrease in expression in vivo compared with in vitro growth (see Tables 4 and 5 and Data Set 3, which are published as supporting information on the PNAS web site).
We determined the functional categories of the in vivo-induced and -repressed genes (Fig. 2B). Except for the group of unknown, hypothetical, and conserved hypothetical proteins, the largest group of genes showing increased expression in vivo was the transporter and binding protein class (Fig. 2B). This group contained 52 in vivo-induced genes (22.6% of the 230 in vivo-induced genes), suggesting that V. cholerae enhances expression of these genes to transport materials that are critical for in vivo survival (e.g., iron). Consistent with an increase in anaerobic energy metabolism in vivo, we observed enhanced expression of 13 genes involved in anaerobic carbohydrate metabolism (e.g., glpABC, frdABCD, pflB, adhE, and oadA) and four genes (dcuABC and dctP) involved in anaerobic carbohydrate transport. Genes encoding regulatory proteins accounted for 26 of the 230 in vivo-induced genes (Fig. 2B), indicating major shifts in the metabolism and cell biology of V. cholerae in vivo vs. in vitro.
There are 12 pathogenesis genes among the 230 genes classified as in vivo-induced (Fig. 2B): mshABCDE, tcpPH, hlyAB, hap, irgB, and toxS. Seven of these were among the 300 most highly expressed genes. As noted above, mshABCD encode a type IV pilus involved in adhesion to mucosal receptors, and it is also a known target of the human immune response (26, 27). TcpPH and ToxS are early members of a regulatory cascade that ultimately leads to expression of TCP and CT (4). Motility is also a virulence phenotype of V. cholerae (28), and 10 genes in this group were in vivo-induced. In contrast, no flagellar or chemotaxis genes were among the 296 in vivo-repressed genes. These data suggest that V. cholerae enhances its virulence properties on entering the host by increasing expression of colonization, motility and chemotaxis genes.
Of the 296 genes with decreased expression in vivo, 81 (27.4%) and 39 (13.2%) are involved in protein synthesis and energy metabolism, respectively (Fig. 2B). Interestingly, many of these genes are among the 300 most highly expressed genes in ileal loops, indicating that protein synthesis and energy metabolism proceed during growth in vivo but at a much lower rate than in LB.
Discussion
Understanding bacterial gene expression patterns during the pathogen–host interaction has long been a goal of investigators interested in pathogenesis, bacterial physiology, and the host immune response to infectious agents. An appreciation of the limitations of studying surrogate in vitro signals as the cues for controlling gene expression has led the field to explore genetic approaches that could define “in vivo-induced genes” for pathogens (29–31). However, in vivo induction is an arbitrary parameter to measure because assumptions must be made about which in vitro condition is appropriate for comparison to any in vivo condition. In this study, we attempted to measure gene expression in vivo and in vitro during comparable growth phases.
We compared the genomic transcriptional pattern (i.e., the transcriptome) of in vivo-grown cells with that of cells grown aerobically under laboratory conditions. Under both conditions, the genes showing the highest levels of expression reside primarily on the V. cholerae large chromosome. However, many more small-chromosome genes were expressed during in vivo growth. Our analysis suggests that iron limitation, anaerobiosis, and nutrient limitation are prominent environmental conditions encountered by V. cholerae during growth in the rabbit upper intestine. For example, the expression of 24 or more genes involved in iron transport or storage was increased in vivo. Enhanced expression in vivo was observed for 13 anaerobic energy metabolism genes, including the regulatory gene arcA that is known in other enteric organisms to be anaerobically induced and required for repression of many aerobic metabolism operons (32). These data suggest that V. cholerae in ileal loops may actively scavenge organic molecules such as fumarate, formate, and glycerol-3-phosphate (all involved in anaerobic respiration), maltose and fructose, and peptides/amino acids. Finally, other genes showing high expression (e.g., bioC, bioD, and bisZ) indicate that vitamins such as biotin may be unavailable in the intestine. This result correlates with signature-tagged mutagenesis (STM) studies where V. cholerae biotin biosynthesis mutants were shown to be defective in intraintestinal growth (6). In fact, 11 genes scored in our study as in vivo-induced in ileal loops were previously found by STM to be required for intestinal colonization in suckling mice (sspA, tonB1, rfbD, rfbE, cycA, tcpP, frdC, ackA, pta, nqrD, and nqrC) (6, 7).
Relative to in vitro conditions, the intestinal environment significantly enhanced expression of many virulence genes, including those involved in adherence, motility/chemotaxis, and regulation of TCP and CT expression. For example, TcpPH and ToxS (all induced in ileal loops) function early in the regulatory cascade controlling TCP and CT expression (4). The genes for MSHA pili were also induced in ileal loops, and these pili are thought to mediate binding to mucosal receptors (27). Finally, 10 genes involved in motility and chemotaxis were significantly induced, consistent with this behavior playing a significant role in early intestinal colonization events (28). This is a modest number given that >100 V. cholerae genes are annotated as functioning in chemotaxis or motility, but we believe it is significant because no genes of this sort were found to be repressed in ileal loops.
Recently, Merrell et al. found 44 induced and 193 repressed genes in V. cholerae cells shed from cholera patients compared with V. cholerae grown to stationary phase in LB (12). However, only 3 of the 44 in vivo-induced genes (encoding a formate transporter, cold-shock domain family protein, and a multidrug resistance protein) are present on our list of genes induced in rabbit ileal loops compared with growth in exponential phase in LB. Clearly, more work on expression profiling of V. cholerae derived from cholera clinical samples is needed to fully understand these divergent results.
Although CT and TCP are important virulence determinants (1, 3, 33), only tcpP, tcpH, and toxS were found to be induced in vivo; however, toxR was among the top 300 genes expressed in vivo, although it was not induced in vivo. Merrell et al. also found no differential expression of genes in the ToxRS/TcpPH/ToxT regulon in V. cholerae present in patient stools (12). However, we observed that 5- to 10-fold more fluid accumulated in rabbit ileal loops inoculated with V. cholerae than loops inoculated only with PBS; this fluid accumulation indicates that CT was likely expressed by V. cholerae at 8 h postinoculation. Perhaps CT expression occurs either transiently or only in a minority of the in vivo population (e.g., only in cells adhering to the intestinal epithelium). In other experiments, we actually observed decreased tcpPH expression in V. cholerae recovered from ileal loops where vibrios grew to much higher density (109–1010 cells/ml) and thus were presumably in stationary phase in vivo (Q.X. and J.J.M., unpublished data). In general, we have observed that gene expression patterns in these “stationary-phase” in vivo-grown cells differ dramatically from those from the midexponential-phase cells harvested from the loops. For example, among the 300 most highly expressed genes in stationary phase, in vivo-grown cells, only 18 encode ribosomal proteins or translation factors (compared with 71 such genes for midexponential phase in vivo-grown cells). Furthermore, “stationary-phase” cells harvested from rabbit loops show an even more pronounced transcriptional activation of genes on the small chromosome (Q.X. and J.J.M., unpublished data). Thus, we conclude that gene expression patterns of V. cholerae during different stages of in vivo growth will be different.
The two-chromosome structure of the V. cholerae genome is common to many other Vibrio species (34). Our work suggests that genes on the small chromosome of V. cholerae are activated in expression during intraintestinal growth. We propose that the activation of small-chromosome genes at all stages of growth in vivo probably reflects a specific role for the small chromosome in the response to unique nutritional stresses within the host environment. The small chromosome of other Vibrio species may play a similar role with alternative aquatic hosts (e.g., shellfish, squid, fish, aquatic mammals, copepods, etc.). Our proposed role for the small chromosome in adaptation to “host-related” nutritional stresses may partly explain the phenomenal success of the Vibrio genus as environmental organisms.
Supplementary Material
Acknowledgments
We thank Emmy Balon for technical assistance in the construction of V. cholerae microarrays, Daniele Provanzano for his assistance in rabbit experiments, Dan Fraenkel for helpful discussions, and Su Chiang for critically reading and editing the manuscript. This work was funded by National Institutes of Health Grant AI18045 (to J.J.M.).
Abbreviations
- CT
cholera toxin
- TCP
toxin-coregulated pilus
- gDNA
genomic DNA
References
- 1.Faruque S M, Albert M J, Mekalanos J J. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waldor M K, Mekalanos J J. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 3.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotter P A, DiRita V J. Annu Rev Microbiol. 2000;54:519–565. doi: 10.1146/annurev.micro.54.1.519. [DOI] [PubMed] [Google Scholar]
- 5.Lee S H, Hava D L, Waldor M K, Camilli A. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 6.Chiang S L, Mekalanos J J. Mol Microbiol. 1998;27:797–806. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 7.Merrell D S, Hava D L, Camilli A. Mol Microbiol. 2002;43:1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- 8.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Judson N, Mekalanos J J. Nat Biotechnol. 2000;18:740–745. doi: 10.1038/77305. [DOI] [PubMed] [Google Scholar]
- 10.Dziejman M, Balon E, Boyd D, Fraser C M, Heidelberg J F, Mekalanos J J. Proc Natl Acad Sci USA. 2002;99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J, Miller M B, Vance R E, Dziejman M, Bassler B L, Mekalanos J J. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merrell D S, Butler S M, Qadri F, Dolganov N A, Alam A, Cohen M B, Calderwood S B, Schoolnik G K, Camilli A. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De S H, Chatterjee D N. J Pathol Bacteriol. 1953;46:559–562. doi: 10.1002/path.1700660228. [DOI] [PubMed] [Google Scholar]
- 14.Talaat A M, Howard S T, Hale W T, Lyons R, Garner H, Johnston S A. Nucleic Acids Res. 2002;30:e104. doi: 10.1093/nar/gnf103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudley A M, Aach J, Steffen M A, Church G M. Proc Natl Acad Sci USA. 2002;99:7554–7559. doi: 10.1073/pnas.112683499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Y, Lee J M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tendeng C, Badaut C, Krin E, Gounon P, Ngo S, Danchin A, Rimsky S, Bertin P. J Bacteriol. 2000;182:2026–2032. doi: 10.1128/jb.182.7.2026-2032.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waldor M K, Mekalanos J J. Infect Immun. 1994;62:72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller V L, Mekalanos J J. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golby P, Kelly D J, Guest J R, Andrews S C. J Bacteriol. 1998;180:6586–6596. doi: 10.1128/jb.180.24.6586-6596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Six S, Andrews S C, Unden G, Guest J R. J Bacteriol. 1994;176:6470–6478. doi: 10.1128/jb.176.21.6470-6478.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyckoff E E, Stoebner J A, Reed K E, Payne S M. J Bacteriol. 1997;179:7055–7062. doi: 10.1128/jb.179.22.7055-7062.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sciortino C V, Finkelstein R A. Infect Immun. 1983;42:990–996. doi: 10.1128/iai.42.3.990-996.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller V L, Mekalanos J J. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Häse C C, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qadri F, Jonson G, Begum Y A, Wenneras C, Albert M J, Salam M A, Svennerholm A M. Clin Diagn Lab Immunol. 1997;4:429–434. doi: 10.1128/cdli.4.4.429-434.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonson G, Lebens M, Holmgren J. Mol Microbiol. 1994;13:109–118. doi: 10.1111/j.1365-2958.1994.tb00406.x. [DOI] [PubMed] [Google Scholar]
- 28.Richardson K. Infect Immun. 1991;59:2727–2736. doi: 10.1128/iai.59.8.2727-2736.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahan M J, Slauch J M, Mekalanos J J. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 30.Camilli A, Beattie D T, Mekalanos J J. Proc Natl Acad Sci USA. 1994;91:2634–2638. doi: 10.1073/pnas.91.7.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valdivia R H, Falkow S. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 32.Iuchi S, Lin E C. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tagomori K, Iida T, Honda T. J Bacteriol. 2002;184:4351–4358. doi: 10.1128/JB.184.16.4351-4358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.