Fig. 2.
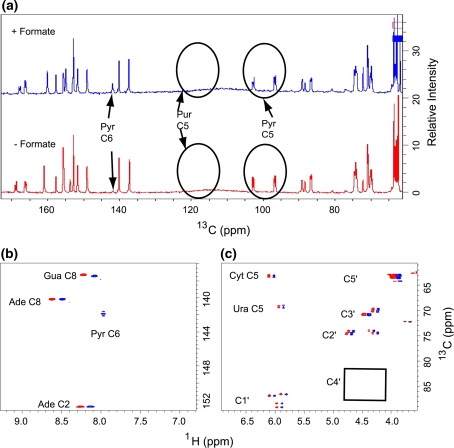
Labeling pattern of a mixture of four rNMPs isolated from DL323 E. coli strain grown without (red) and with (blue) 13C-formate in a 13C-1,3-glycerol background. a Direct carbon detection 1D spectrum showing all the labeled carbon positions for nucleotides labeled with 13C-1,3 -glycerol and no formate (bottom, red) or with formate (top, blue). A long recycle delay of 5 s was used to allow for sufficient magnetization recovery and proton decoupling was limited to the acquisition period only. The level of enrichment at the adenine (Ade) and guanine (Gua) C8 positions remain at the same high level but that of Pyr C6 increases only on addition of 13C-labeled formate. Slight differences in pH and salt conditions between samples leads a noticeable shift in the chemical positions for peaks downfield of Pyr C6. b 2D non-constant time HSQC spectrum of a mixture all four labeled rNMPs showing the protonated base region. For ease of comparison the spectrum obtained without labeled formate (red contours) are displaced to the left of the formate labeled spectrum (blue contours). The level of enrichment at the Pyr C6 increases slightly by spiking with 13C-labeled formate. c 2D non-constant time HSQC spectrum of a mixture of all four labeled nucleotides showing the ribose region. The cytosine (Cyt) and Uracil (Ura) C5 resonances at 96.67 ppm and 102.69 ppm respectively are folded into the spectrum. The C4′ region is boxed to highlight the absence of labeling