Abstract
The use of stem cells for regenerative medicine has captured the imagination of the public, with media attention contributing to rising expectations of clinical benefits. Human embryonic stem cells (hESCs) are the best model for capital investment in stem cell therapy and there is a clear need for their robust genetic characterization before scaling-up cell expansion for that purpose. We have to be certain that the genome of the starting material is stable and normal, but the limited resolution of conventional karyotyping is unable to give us such assurance. Advanced molecular cytogenetic technologies such as array comparative genomic hybridization for identifying chromosomal imbalances, and single nucleotide polymorphism analysis for identifying ethnic background and loss of heterozygosity should be introduced as obligatory diagnostic tests for each newly derived hESC line before it is deposited in national stem cell banks. If this new quality standard becomes a requirement, as we are proposing here, it would facilitate and accelerate the banking process, since end-users would be able to select the most appropriate line for their particular application, thus improving efficiency and streamlining the route to manufacturing therapeutics. The pharmaceutical industry, which may use hESC-derived cells for drug screening, should not ignore their genomic profile as this may risk misinterpretation of results and significant waste of resources.
Keywords: human embryonic stem cells; cell therapy; chromosome instability; karyotype; array comparative genomic hybridization, copy number variations; single nucleotide polymorphism
1. Human embryonic stem cells: the best stem cell model for capital investment in stem cell therapy
Media-fuelled belief that stem cells possess a virtually unlimited restorative power and represent a universal remedy for all diseases has resulted in considerable funding support and scientific progress in stem cell biology over the last decade. However, despite exciting prospects, the potential of stem cells has not yet materialized. Patients with serious degenerative disorders such as Parkinson's disease, diabetes or cardiac disease have been waiting for the fulfilment of these media-hyped promises, but as yet there are a relatively small number of clinical trials despite years of research in this field.
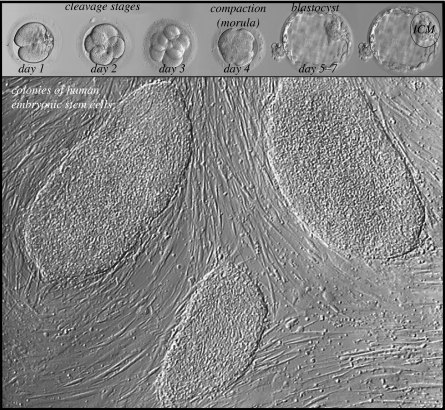
Most commonly, embryonic stem cells (ESCs) are generated from the inner cell mass of the blastocyst (figure 1). In vitro these appear to be immortal, proliferating indefinitely in an undifferentiated, pluripotent state (figure 2). However, ESCs in vivo lose this property as differentiation proceeds and as development and growth-promoting signals change. By adulthood, the few remaining stem cells are dispersed throughout the body and are difficult to locate. However, they seem to be able to continue to generate identical daughter cells and/or tissue cells at each division. These residual pools of stem cells are suggested to be the source of the tissue regeneration and repair that occurs in adults. Tissue-specific stem cells are present in many organs and systems in adult animals although they differ greatly in their ability to self-renew and differentiate. For example, spermatogonial stem cells in the testis are unipotent and produce only one type of differentiated cell—the spermatozoon. By contrast, mesenchymal stem cells are multipotent and can produce adipocytes, osteoblasts, chondrocytes and myocytes in appropriate culture conditions. Unlike their embryonic counterparts, tissue-specific stem cells are not immortal, and show a decreasing capacity to self-renew with increasing age. This limitation has been associated with the reduced ability to repair the damage that accumulates with ageing, possibly owing to exhaustion of the stem cell pool, or as a consequence of inherited or acquired mutations throughout life that impede normal stem cell function.
Figure 1.
Most commonly, human embryonic stem cells are derived from inner cell mass (ICM) of the blastocyst plated on mitotically inactivated fibroblasts.
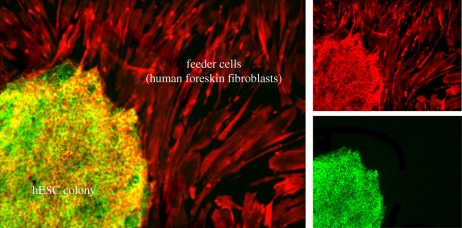
Figure 2.
Human embryonic stem cell (hESC) colony. hESCs have high alkaline phosphatase activity (green). Actin filaments (red) are visualized with rhodamine X-conjugated phalloidin in both hESCs and surrounding human foreskin fibroblast feeders.
The difficulty in locating these scarce stem cells from a variety of sources and expanding their number sufficiently for therapeutic use is proving a major hindrance for industry in the translation of stem cell potential to be used in regenerative medicine. Pluripotent stem cells, both human ESCs (hESCs) and induced pluripotent stem cells (iPSCs), are exceptions since they can be produced in theoretically limitless quantities, and therefore capable of providing more cells than from any other source, regardless of differentiation efficacy and stabilization. Thus, they are the cell type likely to yield the most from invested capital. Although both types of stem cells are promising, there are still a number of unresolved technical and biological issues that make iPSCs less likely to be the immediate choice for cell-based therapy. This leaves the pluripotent hESCs as the best stem cell model for capital investment for stem cell therapy.
The initial difficulties with legislation to allow use of human embryos for stem cell research, the lack of consensus for reporting the quality and type of embryos suitable for stem cell derivation, and the route to translation have largely been overcome, especially in the UK, where a regulatory route map to facilitate clinical application has recently been produced as a collaborative project between the regulatory bodies involved: Human Fertilisation and Embryology Authority (HFEA), Human Tissue Authority (HTA) and Medicines and Healthcare Products Regulatory Agency (MHRA).
2. Risk assessment of genetic abnormalities in hesc-based cell therapy
Although reliable and validated methods for deriving, growing and preserving hESCs are within reach, the greatest impediment to the provision of hESCs for therapeutic purposes is that full genotypic and phenotypic characterization criteria have not yet been established, on a scale that can be routinely and reliably applied. Despite wide interest in defining the properties of hESCs, comprehensive characterization has been done with only a subset of lines. The popularity of some of the earliest lines derived, such as H1 and H9, both for research and for the generation of clinical grade banks, arises not necessarily because they are superior lines, as these cells were derived on mouse feeders in the presence of serum, but because by default they are the best characterized. Although variations in derivation and culture conditions can account for differences among lines, even those derived under identical conditions do differ. This genotypic instability represents one of the most critical hurdles in the development of hESC lines as therapeutic products, in terms of cell-based therapy as well as high-throughput drug screening, and a risk for investors considering commercialization opportunities.
The gene-expression profile of hESCs explored by sophisticated techniques, such as serial analysis of gene expression, expressed sequence tag enumeration, microarray analysis and massively parallel signature sequencing, has revealed surprisingly large variations among lines (Adewumi et al. 2007; Allegrucci & Young 2007; Lefort et al. 2008; Närvä et al. 2010). Although many of such studies have been useful in identifying ‘master’ genes of pluripotency, no consistent clustering could be ascribed to variations in chromosomal stability or differentiation propensity of the hESC lines. The number of differences detected is likely to reflect random fluctuations, and no approach has emerged to obviate the establishment of a cell line signature. Both inherent instability within the embryos used to derive the lines, or selection pressures owing to culture conditions could contribute to this variation. To define what is an acceptable cell signature, extensive high-resolution genomic analyses should be done on a large number of hESC lines at early and late passages, cultured under various conditions. Only meta-analysis of such studies might lead to a conclusion.
3. Embryos used for hesc derivation
From 2004, the Medical Research Council awarded a number of strategic grants to UK centres in an effort to facilitate stem cell research. These grants not only provided funds to set up stem cell laboratories in proximity to in vitro fertilization (IVF) units, but also facilitated collaboration between IVF units that had access to human embryos surplus to therapeutic need and scientists trying to derive stem cells. Since it was a requirement of HFEA licences for stem cell research that a sample of any line derived should be deposited in the UK Stem Cell Bank (UKSCB) for international use in research, it also complied with the spirit of the HFEA Act, whereby the number of embryos used for research should be limited by necessity. A uniform patient information and consent process was introduced with the formation of the hESC coordinators network (hESCCO; Franklin et al. 2008). The requirement to have an arms-length trained person to talk to interested patients about the research not only ensured rigour of informed consent, but also streamlined the process and facilitated national collaboration between centres to increase the number of embryos available for stem cell research.
These goals have largely been realized with five UK centres currently attempting to derive hESC lines within a GMP (good manufacturing practice) environment. If cells are intended for human use, the premises and processes are required to conform to the Human Tissue Act (2004) and thus be licensed by the HTA, and also to conform to practices approved by the MHRA. These require the basic information about the donors of the sperm and eggs, in order to minimize risks of transmission of adventitious agents. The European Directive on Cells and Tissues also requires that there should be basic relevant information from the patients' histories, and as a minimum testing for human immunodeficiency virus (HIV) 1 and 2, hepatitis B and C, and syphilis. It is argued that requiring more detailed personal information as is required for blood donors would not only be intrusive but also likely to be unreliable as the ‘donors’ are seen together and may be unlikely to reveal truthful information about the previous or present sexual environment. Similarly, it is neither easy nor necessarily practical to return to the donor couple if further information or tests related to stem cells are needed—these couples are ‘incidental’ donors, as their primary reason for attending IVF is to have a baby. Additionally, as only a minute proportion of embryos generated for IVF will ever be used for stem cell derivation, it is argued that retaining a sample of serum from each patient for later testing would be impractical. Stem cell lines are unique among tissues for donation and therapy in that they can be amplified and stored before release, thus allowing additional post hoc testing for safety. Although one can never ‘test safety into a product’, the availability of large amounts of material and development of new in vitro tests should substantially reduce risk, and be better than inference from unreliable medical histories, or limited testing on pre-derivation serum samples.
4. Inherited and acquired structural chromosomal abnormalities
Chromosomal instability has been reported for several widely used hESC lines (H1, H7, H14, HS181, HS237, SA002.5, hESC5 and BG01—e.g. Hanson & Caisander 2005; Allegrucci & Young 2007; Baker et al. 2007; Catalina et al. 2008). These karyotypic changes emerged beyond passage 13 and were in general losses or gains of whole chromosomes (aneuploidy), rather than structural rearrangements within a diploid karyotype. However, other studies reported a lack of karyotypic changes in a variety of hESC lines (SA001, hES1-6, BG02, BG03, SA003, SA121, SA461, HS235) even when grown for between 34 and 140 passages. Although this cytogenetic resilience of some hESC lines might arise from particular aspects of cell culture (passage methods, presence versus absence of feeders, and so on), it might also reflect an inherent genetic predisposition of some hESC lines to chromosomal instability. In either event, in order to use them as models, there is a legitimate need to identify patient/lineage-specific properties in the starting material and any pathological phenotypes in disease-specific pluripotent cell lines.
4.1. Intrinsic factors—inherent instability
hESC lines are derived from embryos made available following IVF for the treatment of female and male infertility, or from preimplantation genetic diagnosis (PGD) treatment cycles undertaken to avoid transmitting a known genetic disease. Physical or hormonal problems may explain some causes of infertility, but in many cases the reason why a couple is unable to conceive remains unknown; one explanation may be the genetic status of the embryos. Chromosomal abnormality is common in human embryos. Over 50 per cent of preimplantation IVF embryos have chromosomal abnormalities such as aneuploidy, polyploidy and haploidy (Munné et al. 1994; Harper et al. 1995; Marquez et al. 2000; Ilic et al. 2010). Most of these abnormalities are not compatible with embryo development. This is one explanation for why under half of all human conceptions result in live births (Edmonds et al. 1982) and for the relatively low implantation rate after IVF, even when morphologically high-quality embryos are transferred (Gianaroli et al. 1999; Kahraman et al. 2000; Voullaire et al. 2007). It might also be reasonable to speculate that some infertility might be caused by undiagnosed genetic mutations, which affect either embryo quality or the implantation process. Thus, an embryo created by IVF, and consequently the hESC lines derived from it, might also be affected by such abnormalities or mutations. Although genetic links between impaired fertility and susceptibility to various degenerative diseases are yet to be established, the prevalence of balanced structural chromosomal abnormalities in patients seeking IVF or PGD treatment is higher than in the general population. This further highlights the need for comprehensive genetic analyses to become a routine part of hESC line characterization. Alterations at the chromosome level might result in undesirable consequences for the recipients of stem cell therapies (Amariglio et al. 2009).
4.2. Extrinsic factors
Normal human cells in vivo have a rate of spontaneous mutation of 10−7 to 10−8 per nucleotide per cell division. Since there are approximately 3 × 109 nucleotides per haploid human genome, between 30 and 3000 mutations could occur per cell at each cell cycle (Lefort et al. 2009). Fidelity of DNA replication in vitro is likely to be even lower.
The predominant spontaneous mutation in ESCs is the loss of heterozygosity (LOH; Cervantes et al. 2002). In somatic cells, LOH is mediated mostly by mitotic recombination, which is suppressed in ESCs. Chromosome loss/reduplication leading to uniparental disomy (UPD) represents more than half of the LOH events in ESCs, a pathway that is not commonly observed in other somatic cells. Culture parameters and manipulation techniques have been shown to have an effect on gene-expression patterns and genomic integrity in both embryos and stem cells. For example, the hESC lines H1 and H14 grown in a laboratory at the University of Wisconsin predominantly gained an extra chromosome 12, whereas at the University of Sheffield the same lines gained an extra chromosome 17 (Werbowetski-Ogilvie et al. 2009).
4.2.1. Oxygen tension
Both preimplantation embryos from which hESC lines are derived and hESC lines themselves are particularly sensitive to the oxygen tension to which they are exposed in vitro. It is well known that an increase in free oxygen radicals has an adverse effect on fertilization rates, embryo quality and growth in vitro. However, even though hESCs have been shown to grow as well under 3 or 5 per cent oxygen as at 21 per cent, corroborating results have shown reduced spontaneous differentiation, enhanced cell proliferation, and increased plating, freezing and thawing efficiency after maintenance in low-oxygen tension for more than 14 passages (Ezashi et al. 2005). Furthermore, oxygen tension of 2 per cent has been shown to enhance hESC clonal recovery and significantly reduce the acquisition of spontaneous chromosomal abnormalities (Forsyth et al. 2006). Since hESCs have metabolic characteristics of preimplantation embryos, which must rely on anaerobic metabolism instead of oxidative phosphorylation to produce adenosine-5′-triphosphate at 1–5% oxygen tension in the uterus, they have fewer and smaller mitochondria with poorer cristae (Oh et al. 2005; St John et al. 2005). This indicates that the hESCs may not be as well protected against DNA damage caused by reactive oxygen species and free radicals, which are generated during normal oxidative cell metabolism. Indeed, undifferentiated hESCs have a much lower expression of Cu/Zn-superoxide dismutase, glutathione peroxidase 1, and peroxiredoxin 1 and 2, which are enzymes that normally protect cells against such damage (Cho et al. 2006).
4.2.2. Use of enzymes for disaggregating colonies
Several groups have reported that the technique used for passaging cells can induce alterations in karyotype (Draper et al. 2004; Hoffman & Carpenter 2005; Mitalipova et al. 2005). Comparison of mechanical cutting of colonies and the use of enzymes for passage in the same hESC line suggested that mechanical cutting better supports maintenance of a normal karyotype in long-term culture; the enzyme-treated cell line was more likely to develop karyotype abnormalities (most often chromosome 12 and 17 trisomy). The type of enzyme used does not seem to play a role; the phenomenon is observed with all three commonly used enzymes for hESC passage: collagenase, trypsin or accutase. Although it is unclear why the enzyme-based methods favour gross chromosomal rearrangements when this is not seen with other cell types such as fibroblasts, which are passaged in a similar way in vitro, the most likely explanation is that it is linked to well-established dynamic reciprocity between architectural integrity through cell adhesion and karyotype stability (Tlsty 1998). hESCs are polarized and express an epithelial plasma membrane protein profile (Krtolica et al. 2007; Van Hoof et al. 2008). Frequent disruption of cell–cell contacts and polarity may induce karyotype rearrangements and DNA damage that would, in differentiated cells, lead to cell death. However, in order to favour cell proliferation as in early embryos in vivo, DNA-repair mechanisms that protect the genome from endogenous and exogenous factors that induce DNA damage and other genotoxic insults, are not fully developed in ESCs. The activity of genes regulating chromosome segregation, the cell cycle and apoptosis fluctuate during human preimplantation development (Wells et al. 2005). The guardian of the genome, the tumour suppressor gene, p53, which prevents the accumulation of genetic mutations in somatic cells by inducing cell cycle arrest, apoptosis or senescence, does not function in ESCs in response to DNA damage in the same way as in somatic cells. Instead of p53-dependent apoptosis or cell cycle arrest at the G1/S checkpoint following DNA damage, in ESCs p53 suppresses nanog expression and pushes the cells towards differentiation (Aladjem et al. 1998; Song et al. 2010; Zhao & Xu 2010).
Therefore, in order to avoid gross abnormalities, it seems that hESC lines are best passaged manually using mechanical splitting until karyotypically normal frozen stocks are established. However, even manual passage does not protect against structural rearrangements such as microdeletions or duplications. For example, according to fluorescent in situ hybridization analysis, an amplification of 2.5–4.6 Mb at 20q11.21, encompassing 23 genes, has been reported in the oldest hESC line H1 already at passage 24 (Lefort et al. 2008). The number of rearrangements was relatively high at passage 56, even though the passaging has been done manually (Hovatta et al. 2010). However, it is possible that these imbalances had been present from the beginning, since no analyses with such a resolution had been done on earlier passages. Sub-populations of hESCs that accumulate karyotype alterations usually have a growth advantage. Occasionally, though unfortunately not always, they can be selected against when passaged mechanically and the operator is able to make the decision about splitting based on morphological criteria. Before scaling-up cell expansion for the purpose of cell therapy, we have to be certain that the genome of our starting material is stable and normal. The resolution of G-banded karyotype is unable to give such assurance.
4.2.3. Feeder-free culture
In one study, which looked at parameters of functional adaptation to growth directly on a plastic surface during long-term culture of hESCs, successful adaptation was achieved but was paralleled with a karyotype change in 100 per cent of the cells (Imreh et al. 2006). Similarly, initial attempts at derivation in feeder-free-defined conditions resulted in hESC lines with unstable karyotypes (Ludwig et al. 2006). A microenvironment, which retained a three-dimensional extracellular matrix, fared much better in the maintenance of normal karyotype (Ilic 2006). Matrigel, a solubilized basement membrane preparation extracted from the Engelbreth-Holm-Swarm mouse sarcoma, a tumour rich in extracellular matrix proteins, supports large-scale propagation of hESCs without leading to chromosomal imbalances, especially if the cells are passaged mechanically (Xu et al. 2001). This reinforces the importance of the dynamic reciprocity between cell adhesion and karyotype stability as discussed above.
4.2.4. Cryopreservation
The repeated freeze–thaw cycles, whether by conventional slow-cooling methods or vitrification, are unavoidable when banking and expanding cells. Cryo-damage occurs owing to the formation of orderly ice-crystalline lattice structures. Although hESCs are infamous for relatively low recovery upon thawing, the extent to which cryopreservation affects hESC genomic integrity is unknown. The most widely used cryoprotectant dimethylsulphoxide modifies the epigenetic profile of ESCs and increases the production of free radicals that can affect DNA replication and chromatin structure (Iwatani et al. 2006; Diaferia et al. 2008). Ice crystals and gas bubbles formed during thawing can disrupt spindles and induce abnormal segregation of chromosomes (Diaferia et al. 2008). Evidence from zebrafish studies suggests that cryopreservation increases the frequency of mutations in the mitochondrial genome by nearly fivefold (Kopeika et al. 2005). If the normal rate of spontaneous mutations is between 30 and 3000 per cell during each cell cycle, this finding suggests that cryopreservation may result in 150–15 000 mutations in each surviving cell and needs a sufficiently sensitive system to detect them.
5. Current criteria for hesc line acceptance into stem cell banks
Lack of applied technology has resulted in the deposition of most hESC lines in both UK and USA stem cell banks with no more than basic information such as G-banded karyotype and marker expression. This is a cause for the anxiety of manufacturers and end-users and is one factor that may have prevented hESC utilization at the rapid pace that befits their enormous potential. Since such an enormous investment is needed to take a cell line through to a medicinal product, manufacturers need to know as much as possible about the available cell lines in order to select the one most likely to be suitable for developmental needs.
For centres deriving hESC lines in the UK, it is a condition of the HFEA licence that a sample must be deposited in the UKSCB. Therefore, the UKSCB cannot enforce a minimum requirement for characterization of research grade lines, but can make a decision as to which to process further or which to simply archive. However, to deposit a line to be declared of clinical grade, a minimum characterization requirement needs to be established and is being considered to include passage number before submission, DNA fingerprinting, karyotype, viral and sterility testing, viability assays and expression of pluripotent markers.
Advantages cited for the inclusion of cell lines in an international bank, such as the UKSCB, include promotion of the wider use of the cell line by providing stringently tested, well-characterized cells within a quality framework, detailed assessment using standardized methodologies and optimized culture, preservation and characterization through a targeted research programme (www.ukstemcellbank.org.uk). In order for this mission to be achieved, cells will need to be supplied to the UKSCB with as much information as possible, which should include a detailed genotype. In the report describing their derivation of some of the few clinical grade stem cell lines that exist, the A*Star Singapore team provide no more than routine characterization (G-banded karyotype and pluripotent/differentiation marker expression by immunocytochemistry, polymerase chain reaction and fluorescence activated cell sorting), which seems insufficient for confident selection (Crook et al. 2007). There is now a legitimate need to identify lineage-specific properties in the starting material using modern molecular methods for every newly derived hESC line before the line is deposited in national stem cell banks in order to use hESCs as a source in cell-based therapy.
6. Molecular karyotyping
6.1. Copy number variations: molecular diagnostics
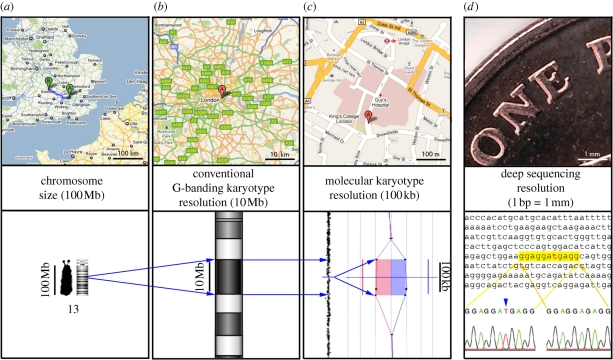
For the last 50 years, karyotype analysis has been the ‘gold standard’ for detection of chromosome anomalies. From plain staining and counting, to extended chromosomes, enzymatically treated and stained to give detailed band-by-band comparison along homologous pairs, the methodology for producing chromosome preparations has been improved over the years (Yunis 1976). However, even with optimal conditions, the resolution of this technique is considered to be approximately 3 Mb, while in certain regions of the genome, imbalances of 10 Mb can be hard to detect. G-banded karyotype analysis requires lengthy training for the operatives, and is very subjective, depending on highly developed pattern recognition skills in the analyst. In addition, in shorter chromosome preparations, such as those typically obtained from cell lines, only abnormalities of whole chromosome copy number and very large imbalances can confidently be assigned (figure 3).
Figure 3.
Scale illustration of genomic analyses: one-penny sterling coin (diameter 20.32 mm) lying on the road from London (A) to Oxford (B), which is about 100 km, corresponds to 20 bp on human chromosome 13 (100 million bp). The smallest human chromosome (Y) spans about 51 million bp (51 Mb), whereas the largest (chromosome 1) has about 279 million bp (279 Mb) (International Human Genome Sequencing Consortium 2001; Venter et al. 2001). The human chromosome 13 spans about 100 million bp (100 Mb). An average resolution of conventional Giemsa-stained karyotype (G-banding) is 5–10 Mb, whereas molecular karyotyping has a much higher resolution, averaging around 100 kb, which depends on the number of probes spotted on the arrays. (a) 100 Mb on chromosomes corresponds to 100 km on the map. (b) 10 km on the map corresponds to 10 Mb (G-bending resolution). (c) 100 m on the map corresponds to 100 kb (molecular karyotyping resolution). (d) One-penny image: deep-sequencing resolution can distinguish a single basepair mutation (1 mm corresponds to 1 bp).
Advances in molecular cytogenetic technologies have recently led to the introduction of array comparative genomic hybridization (CGH) as either an add-on test following karyotype analysis for the detection of chromosome imbalance (Edelmann & Hirschhorn 2009) or, in an increasing number of centres, as a first-line test (Ahn et al. 2010). This rapid replacement of karyotype analysis by array CGH testing is fuelled by the greatly increased resolution of array CGH, and the objectivity and ease of the analytical process. Various different array CGH platforms are commercially available; most diagnostic laboratories are in agreement that platforms comprising oligonucleotide probes have better resolution and provide more accurate and reproducible results than those comprising bacterial artificial chromosome clones ( Ylstra et al. 2006; Ahn et al. 2010). Oligonucleotide array platforms are available in a number of formats, with different probe densities across the genome. The resolution of the test will therefore depend on the platform of choice. In our experience, an oligonucleotide platform with 44 000 probes can detect regions of imbalance down to approximately 25 kb, a very considerable improvement over the resolution of karyotype analysis (figure 3).
Research studies over the last few years on populations of individuals with no clinical abnormalities have revealed that copy number variation (CNV) in the genome is extremely widespread, with many ‘normal’ people carrying at least one CNV (Iafrate et al. 2004; Feuk et al. 2006). Data on CNVs present in individuals with clinically abnormal phenotypes are collected by a number of central resources such as DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER; https://decipher.sanger.ac.uk/application/) and International Standard Cytogenomic Array (ISCA) Consortium based at the Emory University in Atlanta, Georgia (https://isca.genetics.emory.edu/iscaBrowser/). The ISCA database is complementing benign CNV variants in population, which are collected in the Database of Genomic Variants hosted by the Hospital for Sick Children in Toronto. Recently, the Center for Applied Genomics at the Children's Hospital of Philadelphia has introduced a database of CNVs found in healthy population with normal phenotypes. Information on the clinical significance of CNVs present in individuals with clinically abnormal phenotypes, and their potential relevance to, for instance, complex disease susceptibility, is therefore still emerging.
Online resources have been developed to aid in the interpretation of CNVs; likely pathogenicity can be assessed by such factors as size and gene content. However, small regions containing no known genes cannot necessarily be dismissed as benign, owing to the possibility of position effects on nearby genes, or the presence of controlling factors. Pasting the basepair coordinates of any CNV into a resource such as the Database of Genomic Variants (Iafrate et al. 2004) allows visualization of the region, including all the known genes therein; tracks showing features of genomic architecture can be activated if required. Any published studies showing imbalance for the region in control populations can also be visualized. In addition, click-through links for information on each gene, and links providing information on syndromes and important disease genes can be used for additional interpretation of the likely effect of the imbalance.
Many CNVs are inherited from disease-free parents, but the possibility of effects of genetic background and environmental factors on the expression of phenotype means that these inherited CNVs cannot necessarily be dismissed as benign. De novo benign and pathological CNVs are likely to arise for the most part at meiosis or in very early development, as mosaicism for these CNVs is quite rare. However, such mosaicism has been found (Ballif et al. 2006), indicating that post-zygotic events can give rise to de novo CNVs; nevertheless, CNVs seem to be generally stable in mature lineages.
Characterization of hESC lines prior to deposition in central cell banks is essential. Thus far, G-banded karyotype analysis has been the method of choice for establishing the presence or absence of chromosome anomalies in these cell lines. However, chromosome preparations from these cell lines are notoriously hard to generate, resulting in much time-consuming and in many cases ultimately fruitless work. When successful, the quality of the preparations is almost invariably poor, with short, highly condensed chromosomes, allowing only very low resolution analysis. It seems clear that the improved technology now in place in cytogenetic laboratories should be used to improve the detail and accuracy of the characterization of stem cells. Array CGH investigation of human stem cell lines has already been described by some groups (Xu et al. 2001; Wu et al. 2008; Elliott et al. in press). Array CGH testing of primary cultures, with follow-up testing if required of different passages, would give an overall picture of the CNVs present, and track any changes in the CNV profile engendered by the culture conditions, and/or any differentiation protocols used. Obviously, this testing would not only detect submicroscopic CNVs, but would also detect large chromosome imbalances sometimes found in long-term culture, such as whole chromosome trisomies.
With the introduction of array CGH testing into routine diagnostic cytogenetics laboratories, the expertise and technology for efficient and accurate analysis is widely available. Platforms in use in different laboratories will vary, but providing that the platforms, control DNA source, analysis algorithms and genome build used for any test are fully documented with the CNV profile for each cell line, then cross-platform comparisons will be valid, and the gene content of any CNV can be accessed from online resources (see above). Medium-throughput batch testing of patient diagnostic samples means that stem cell analysis could generally be incorporated with minimum effect on work-flow. Using the resources described above, documentation of gene content and interpretation of the CNVs found could be carried out by the array CGH laboratory, the stem cell laboratory and/or any investigators wishing to use the banked stem cells.
The consumable and hardware costs associated with array CGH testing are considerably higher than for karyotype analysis. However, these costs are falling, driven by commercial competition, advances in the technology of array manufacture and roll-out of testing, leading to larger orders. Conversely, array CGH labour costs are minimal when compared with culture and chromosome preparation and analysis from stem cell lines; labour costs for culture and karyotype analysis are unlikely to fall. There are currently no standardized overall costs for either karyotype analysis or array CGH testing; these costs are highly dependent on local conditions at testing laboratories. However, by using parsimonious hybridization strategies, robotics and batch testing, some laboratories are already able to offer array CGH testing at the same price as karyotype analysis (Ahn et al. 2010). Sending stem cell samples to an existing centre that carries out routine array CGH analysis is therefore a cost-effective and efficient way of collecting the information necessary for proper characterization of genetic imbalance in these cell lines.
6.2. Single nucleotide polymorphism
Single nucleotide polymorphisms (SNPs) are stable substitutions of a single base with a frequency of more than 1 per cent in at least one population. They are located throughout the genome and occur at approximately every 1200 bases. SNP identification has benefited from the availability of sequence data from the human genome project as well as the development of high-throughput SNP genotyping platforms, which allow low-cost whole-genome analysis. SNP genotyping arrays are frequently used in association with linkage studies to locate genes involved in disease. Information on SNP variants is freely available at dbSNP (http://www.ncbi.nlm.nih.gov/snp/; Sherry et al. 2001) with the latest build containing over 23 million refSNPs, of which over 14 million have been validated (Database of Single Nucleotide Polymorphisms (dbSNP), Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine (dbSNP Build ID: 131)).
6.2.1. Ethnic diversity
The International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/index.html.en) was set up to catalogue genetic similarity and variation in humans by comparing genomic sequence in individuals from four populations of African, Asian and European ancestry (International HapMap Consortium 2003). The data show that there is ample evidence of SNP allele frequency differences between distinct ethnicities (International HapMap Consortium 2005). The publicly available data have the potential to be used to identify genes affecting health and disease, as well as those that influence response to drug therapies and vaccinations based on an assumption that haplotype frequencies vary between groups with differing vaccine or drug responses. The hope is that this will lead to improved treatments based on likely response to intervention, ultimately leading to a personalized medicine approach. The latest build of dbSNP contains over 14 million validated SNPs in the human genome, but interrogating each of these would be prohibitively expensive for most projects.
The HapMap initiative has enabled the development of a haplotype (i.e. SNPs that tend to be inherited together as a block) map of the human genome with over 3.1 million SNPs (International Hapmap Consortium 2007), and importantly it has identified a subset of informative SNPs, termed tag SNPs, which can uniquely identify the haplotypes. By only genotyping these tag SNPs (estimated to number approximately 500 000), information regarding the surrounding SNPs (or those in linkage disequilibrium) is captured, without the need to genotype every known SNP, but with whole genome coverage. Park et al. (2007) developed an approach to identify ethnically variant SNPs (ESNPs) using data from the HapMap project. The ESNPs are available in the SNP@Ethnos database (http://bioportal.kobic.re.kr/SNPatETHNIC/), which contains over 100 000 variant SNPs that appear uniquely in each ethnic group.
In an era of investment in pharmacogenomics/pharmacogenetics studies, it is essential to have well-characterized hESC lines, which have many possible applications in toxicology and pharmacology, including screening of compounds during drug development prior to clinical trials. A greater understanding of pharmacogenomics will have a major impact on success and offers the possibility of better patient outcomes with the ability to predict which drugs/vaccines are likely to be effective in individuals of particular genotypes for genes affecting drug metabolism. However, potential benefits of hESC lines in research are currently hampered by the lack of ethnic diversity of available lines (Laurent et al. 2010). With accumulating population-specific SNP data available, it is simple to identify the ethnic origin of a given cell line, as recently demonstrated by Laurent et al. (2010) and Mosher et al. (2010) to enable a more targeted approach by the inclusion of hESC lines derived from populations relevant to the study being conducted.
6.2.2. UPD—a form of LOH
With the development of algorithms to interrogate data generated from SNP arrays (Zhao et al. 2004; Nannya et al. 2005), it is possible to determine chromosome copy number, enabling the detection of aneuploidy and other chromosomal anomalies, such as UPD, with a resolution that is intermediate between cytogenetic techniques and DNA sequencing. This might be particularly useful since hESCs in culture have been shown to acquire small chromosomal amplifications or deletions (Maitra et al. 2005; Lefort et al. 2008; Spits et al. 2008; Hovatta et al. 2010; Närvä et al. 2010).
UPD, a condition in which both alleles have originated from a single parent, occurs as heterodisomy and isodisomy (Engel 1980; Robinson 2000). Heterodisomy, when sequences from both homologues from the transmitting parent are present, might cause abnormalities only if the genes within the involved region are subject to genomic imprinting. Although rare, disruptions of normal gene expression owing to UPD are known; for instance, maternal UPD for chromosome 15 gives rise to Prader-Willi syndrome (OMIM 176270), while paternal UPD for the same chromosome leads to Angelman syndrome (OMIM 105830). Isodisomy, when two identical sequences from one parental homologue are present, could allow transmission of recessive mutations from a heterozygous parent and a number of such debilitating genetic conditions have been reported, including cystic fibrosis, haemophilia A, spinal multiple atrophy and various endocrine neoplasias (Robinson 2000). This can arise either following correction of meiotic monosomy (‘monosomy rescue’), leading to whole chromosome isodisomy, or as a consequence of somatic recombination at mitosis, leading to segmental isodisomy.
Very few hESC lines are analysed for the presence of UPD. Though, among 17 lines analysed by Närvä et al. (2010) one, FES 21, had identical q arms on chromosome 16, whereas all others had a heterozygous set of chromosomes.
7. Conclusions
It is well known that some hESC lines are easier to maintain in culture, whereas others readily undergo spontaneous differentiation. These characteristics govern investigators' choice as to which hESC line they will use in their study. From the first derivation of hESCs (Thomson et al. 1998), the Thomson lines H1, H7 and H9 became the most widely used, just because they are easy to maintain in an undifferentiated state. However, ease of propagation is not necessarily a desirable feature as it may be associated with changes in genomic integrity and therefore loss of therapeutic value. Selective growth advantages provided by genetic abnormalities in the cells may favour such easy maintenance. Indeed, using high-resolution CGH analysis, Hovatta et al. (2010) found 71 genes deleted and 1471 duplicated in ‘normal’ hESC line H1 after less than 60 passages. It has also been reported that H9 has chromosomal abnormalities (Lefort et al. 2008; Werbowetski-Ogilvie et al. 2009).
A study comparing early and late passages of 17 hESC lines (Närvä et al. 2010), using high-resolution genetic techniques, found that in extended cultures, on average, 24 per cent LOH and 66 per cent CNV had undergone changes (calculated false-positive estimate for CNV was 12.5%). A new LOH site with an average of 1000 kb was created at the rate of about 1.3 per passage. It appears that LOH had no preference for a particular chromosome; so far, it was detected in all chromosomes except chromosome 21 and Y.
The physiological process of regeneration, where remaining tissues organize themselves to replace a lost body part, has long been recognized in lower vertebrates such as the phenomenon of limb regeneration in the newt (Brockes 1997). The regenerative medicine field aims to translate the tremendous potential of stem cell biology to achieve tissue or organ regeneration in humans. However, if tissue-specific stem cells or hESCs are to be used to treat a wide variety of human diseases, then several formidable challenges are still to be overcome, which include thorough genotyping and phenotyping, well-controlled derivation and differentiation, cell efficacy, immunogenicity, tumourigenicity, appropriate cell-delivery systems, and short- and long-term safety (Civin & Rao 2006; De Sousa et al. 2006; Gruen & Grabel 2006; Skottman & Hovatta 2006; Bongso et al. 2008; Unger et al. 2008).
The importance of complete validation of any stem cells destined for therapy cannot be underestimated. History demonstrates that infection from donated tissue is possible, often unexpected and devastating. The first case report documenting the transmission of HIV during blood transfusion appeared in 1983 (Ammann et al. 1983). Prion diseases can also be transmitted via tissue and blood donation and may be more ubiquitous than we suspect (Llewelyn et al. 2004; Miller 2009). The substantial risks associated with unregulated therapies have been highlighted by the recent report describing a donor-derived brain tumour following transplantation of uncharacterized foetal neural stem cells (Amariglio et al. 2009). Unlike conventional donation where blood or tissue from one donor may be transplanted to two or three patients, the expansion of stem cell cultures could allow a single hESC line to be used for hundreds, if not thousands of patients. The risk of disease transmission from a single donor thus increases exponentially (Braude et al. 2005).
The route to preventing such events is to perform full characterization and screening of cell lines. Besides prudent post hoc testing for adventitious infective agents, genotype is one of the most important characterization parameters. Submicroscopic changes could render a cell line almost useless with respect to manufacturing a medicinal product, as differentiation and efficacy results may become invalid. This would void the commercialization potential and waste any investment used in development of that line. Array CGH for identifying chromosomal imbalance and submicroscopic CNVs should be introduced as a new quality standard, an obligatory diagnostic testing for every newly derived hESC line before the line is deposited in national stem cell banks. SNP testing for the identification of ethnicity and genome-wide LOH would provide valuable additional information.
Footnotes
One contribution to a Theme Supplement ‘Translation and commercialization of regenerative medicines’.
References
- Adewumi O., et al. 2007. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25, 803–816 (doi:10.1038/nbt1318) [DOI] [PubMed] [Google Scholar]
- Ahn J. W., Mann K., Walsh S., Shehab M., Hoang S., Docherty Z., Mohammed S., Mackie Ogilvie C. 2010. Validation and implementation of array comparative genomic hybridisation as a first line test in place of postnatal karyotyping for genome imbalance. Mol. Cytogenet. 3, 9 (doi:10.1186/1755-8166-3-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem M. I., Spike B. T., Rodewald L. W., Hope T. J., Klemm M., Jaenisch R., Wahl G. M. 1998. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 8, 145–155 (doi:10.1016/S0960-9822(98)70061-2) [DOI] [PubMed] [Google Scholar]
- Allegrucci C., Young L. E. 2007. Differences between human embryonic stem cell lines. Hum. Reprod. Update 13, 103–120 (doi:10.1093/humupd/dml041) [DOI] [PubMed] [Google Scholar]
- Amariglio N., et al. 2009. Donor-derived brain tumor following neural stem cell transplantation in an ataxia telangiectasia patient. PLoS Med. 6, e1000029 (doi:10.1371/journal.pmed.1000029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammann A. J., Cowan M. J., Wara D. W., Weintrub P., Dritz S., Goldman H., Perkins H. A. 1983. Acquired immunodeficiency in an infant: possible transmission by means of blood products. Lancet 321, 956–958 (doi:10.1016/S0140-6736(83)92082-2) [DOI] [PubMed] [Google Scholar]
- Baker D. E., Harrison N. J., Maltby E., Smith K., Moore H. D., Shaw P. J., Heath P. R., Holden H., Andrews P. W. 2007. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 25, 207–215 (doi:10.1038/nbt1285) [DOI] [PubMed] [Google Scholar]
- Ballif B. C., Rorem E. A., Sundin K., Lincicum M., Gaskin S., Coppinger J., Kashork C. D., Shaffer L. G., Bejjani B. A. 2006. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am. J. Med. Genet. A 140, 2757–2767 (doi:10.1002/ajmg.a.31539) [DOI] [PubMed] [Google Scholar]
- Bongso A., Fong C. Y., Gauthaman K. 2008. Taking stem cells to the clinic: major challenges. J. Cell. Biochem. 105, 1352–1360 (doi:10.1002/jcb.21957) [DOI] [PubMed] [Google Scholar]
- Braude P., Minger S. L., Warwick R. M. 2005. Stem cell therapy: hope or hype? BMJ 330, 1159–1160 (doi:10.1136/bmj.330.7501.1159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes J. P. 1997. Amphibian limb regeneration: rebuilding a complex structure. Science 276, 81–87 (doi:10.1126/science.276.5309.81) [DOI] [PubMed] [Google Scholar]
- Catalina P., Montes R., Ligero G., Sanchez L., de la Cueva T., Bueno C., Leone P. E., Menendez P. 2008. Human ESCs predisposition to karyotypic instability: is a matter of culture adaptation or differential vulnerability among hESC lines due to inherent properties? Mol. Cancer 7, 76 (doi:10.1186/1476-4598-7-76) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes R. B., Stringer J. R., Shao C., Tischfield J. A., Stambrook P. J. 2002. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc. Natl Acad. Sci. USA 99, 3586–3590 (doi:10.1073/pnas.062527199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. M., Kwon S., Pak Y. K., Seol H. W., Choi Y. M., Park D. J., Park K. S., Lee H. K. 2006. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem. Biophys. Res. Commun. 348, 1472–1478 (doi:10.1016/j.bbrc.2006.08.020) [DOI] [PubMed] [Google Scholar]
- Civin C. I., Rao M. S. 2006. How many human embryonic stem cell lines are sufficient? A US perspective. Stem Cells 24, 800–803 (doi:10.1634/stemcells.2006-0084) [DOI] [PubMed] [Google Scholar]
- Crook J. M., et al. 2007. The generation of six clinical-grade human embryonic stem cell lines. Cell Stem Cell 1, 490–494 (doi:10.1016/j.stem.2007.10.004) [DOI] [PubMed] [Google Scholar]
- De Sousa P. A., Galea G., Turner M. 2006. The road to providing human embryo stem cells for therapeutic use: the UK experience. Reproduction 132, 681–689 (doi:10.1530/rep.1.01080) [DOI] [PubMed] [Google Scholar]
- Diaferia G. R., Dessi S. S., Deblasio P., Biunno I. 2008. Is stem cell chromosomes stability affected by cryopreservation conditions? Cytotechnology 58, 11–16 (doi:10.1007/s10616-008-9163-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper J. S., et al. 2004. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat. Biotechnol. 22, 53–54 (doi:10.1038/nbt922) [DOI] [PubMed] [Google Scholar]
- Edelman L., Hirschhorn K. 2009. Clinical utility of array CGH for the detection of chromosomal imbalances associated with mental retardation and multiple congenital anomalies. Ann. NY Acad. Sci. 1151, 157–166 (doi:10.1111/j.1749-6632.2008.03610.x) [DOI] [PubMed] [Google Scholar]
- Edmonds D. K., Lindsay K. S., Miller J. F., Williamson E., Wood P. J. 1982. Early embryonic mortality in women. Fertil. Steril. 38, 447–453 [PubMed] [Google Scholar]
- Elliott A. M., Hohenstein Elliott K. A., Kammesheidt A. In press High resolution array-CGH characterization of human stem cells using a stem cell focused microarray. Mol. Biotechnol. (doi:10.1007/s12033-010-9294-1) [DOI] [PubMed] [Google Scholar]
- Engel E. 1980. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am. J. Med. Genet. 6, 137–143 (doi:10.1002/ajmg.1320060207) [DOI] [PubMed] [Google Scholar]
- Ezashi T., Das P., Roberts R. M. 2005. Low O2 tensions and the prevention of differentiation of hES cells. Proc. Natl Acad. Sci. USA 102, 4783–4788 (doi:10.1073/pnas.0501283102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L., Carson A. R., Scherer S. W. 2006. Structural variation in the human genome. Nat. Rev. Genet. 7, 85–97 (doi:10.1038/nrg1767) [DOI] [PubMed] [Google Scholar]
- Forsyth N. R., Musio A., Vezzoni P., Simpson A. H., Noble B. S., McWhir J. 2006. Physiologic oxygen enhances human embryonic stem cell clonal recovery and reduces chromosomal abnormalities. Cloning Stem Cells 8, 16–23 (doi:10.1089/clo.2006.8.16) [DOI] [PubMed] [Google Scholar]
- Franklin S. B., Hunt C., Cornwell G., Peddie V., Desousa P., Livie M., Stephenson E. L., Braude P. R. 2008. hESCCO: development of good practice models for hES cell derivation. Regen. Med. 3, 105–116 (doi:10.2217/17460751.3.1.105) [DOI] [PubMed] [Google Scholar]
- Gianaroli L., Magli M. C., Ferraretti A. P., Munné S. 1999. Preimplantation diagnosis for aneuploidies in patients undergoing in vitro fertilization with a poor prognosis: identification of the categories for which it should be proposed. Fertil. Steril. 72, 837–844 (doi:10.1016/S0015-0282(99)00377-5) [DOI] [PubMed] [Google Scholar]
- Gruen L., Grabel L. 2006. Concise review: scientific and ethical roadblocks to human embryonic stem cell therapy. Stem Cells 24, 2162–2169 (doi:10.1634/stemcells.2006-0105) [DOI] [PubMed] [Google Scholar]
- Hanson C., Caisander G. 2005. Human embryonic stem cells and chromosome stability. APMIS 113, 751–755 (doi:10.1111/j.1600-0463.2005.apm_305.x) [DOI] [PubMed] [Google Scholar]
- Harper J. C., Coonen E., Handyside A. H., Winston R. M., Hopman A. H., Delhanty J. D. 1995. Mosaicism of autosomes and sex chromosomes in morphologically normal, monospermic preimplantation human embryos. Prenat. Diagn. 15, 41–49 (doi:10.1002/pd.1970150109) [DOI] [PubMed] [Google Scholar]
- Hoffman L. M., Carpenter M. K. 2005. Characterization and culture of human embryonic stem cells. Nat. Biotechnol. 23, 699–708 (doi:10.1038/nbt1102) [DOI] [PubMed] [Google Scholar]
- Hovatta O., et al. 2010. A teratocarcinoma-like human embryonic stem cell (hESC) line and four hESC lines reveal potentially oncogenic genomic changes. PLoS ONE 5, e10263 (doi:10.1371/journal.pone.0010263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A. J., Feuk L., Rivera M. N., Listewnik M. L., Donahoe P. K., Qi Y., Scherer S. W., Lee C. 2004. Detection of large-scale variation in the human genome. Nat. Genet. 36, 949–951 (doi:10.1038/ng1416) [DOI] [PubMed] [Google Scholar]
- Ilic D. 2006. Culture of human embryonic stem cells and the extracellular matrix microenvironment. Regen. Med. 1, 95–101 (doi:10.2217/17460751.1.1.95) [DOI] [PubMed] [Google Scholar]
- Ilic D., Caceres E., Lu S., Julian P., Foulk R., Krtolica A. 2010. Effect of karyotype on successful human embryonic stem cell derivation. Stem Cells Dev. 19, 39–46 (doi:10.1089/scd.2009.0136) [DOI] [PubMed] [Google Scholar]
- Imreh M. P., et al. 2006. In vitro culture conditions favoring selection of chromosomal abnormalities in human ES cells. J. Cell. Biochem. 99, 508–516 (doi:10.1002/jcb.20897) [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium 2003. The International HapMap Project. Nature 426, 789–796 (doi:10.1038/nature02168) [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium 2005. A haplotype map of the human genome. Nature 437, 1299–1320 (doi:10.1038/nature04226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (doi:10.1038/nature06258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani M., Ikegami K., Kremenska Y., Hattori N., Tanaka S., Yagi S., Shiota K. 2006. Dimethyl sulfoxide has an impact on epigenetic profile in mouse embryoid body. Stem Cells 24, 2549–2556 (doi:10.1634/stemcells.2005-0427) [DOI] [PubMed] [Google Scholar]
- Kahraman S., Bahçe M., Samli H., Imirzalioğlu N., Yakisn K., Cengiz G., Dönmez E. 2000. Healthy births and ongoing pregnancies obtained by preimplantation genetic diagnosis in patients with advanced maternal age and recurrent implantation failure. Hum. Reprod. 15, 2003–2007 (doi:10.1093/humrep/15.9.2003) [DOI] [PubMed] [Google Scholar]
- Kopeika J., Zhang T., Rawson D. M., Elgar G. 2005. Effect of cryopreservation on mitochondrial DNA of zebrafish (Danio rerio) blastomere cells. Mutat. Res. 570, 49–61 (doi:10.1016/j.mrfmmm.2004.09.007) [DOI] [PubMed] [Google Scholar]
- Krtolica A., et al. 2007. Disruption of apical-basal polarity of human embryonic stem cells enhances hematoendothelial differentiation. Stem Cells 25, 2215–2223 (doi:10.1634/stemcells.2007-0230) [DOI] [PubMed] [Google Scholar]
- Laurent L. C., et al. 2010. Restricted ethnic diversity in human embryonic stem cell lines. Nat. Methods 7, 6–7 (doi:10.1038/nmeth0110-06) [DOI] [PubMed] [Google Scholar]
- Lefort N., Feyeux M., Bas C., Féraud O., Bennaceur-Griscelli A., Tachdjian G., Peschanski M., Perrier A. L. 2008. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat. Biotechnol. 26, 1364–1366 (doi:doi:10.1038/nbt.1509) [DOI] [PubMed] [Google Scholar]
- Lefort N., Perrier A. L., Laâbi Y., Varela C., Peschanski M. 2009. Human embryonic stem cells and genomic instability. Regen. Med. 4, 899–909 (doi:10.2217/rme.09.63) [DOI] [PubMed] [Google Scholar]
- Llewelyn C. A., Hewitt P. E., Knight R. S., Amar K., Cousens S., Mackenzie J., Will R. G. 2004. Possible transmission of variant Creutzfeldt-Jakob disease by blood transfusion. Lancet 363, 417–421 (doi:10.1016/S0140-6736(04)15486-X) [DOI] [PubMed] [Google Scholar]
- Ludwig T. E., et al. 2006. Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 24, 185–187 (doi:10.1038/nbt1177) [DOI] [PubMed] [Google Scholar]
- Maitra A., et al. 2005. Genomic alterations in cultured human embryonic stem cells. Nat. Genet. 37, 1099–1103 (doi:10.1038/ng1631) [DOI] [PubMed] [Google Scholar]
- Marquez C., Sandalinas M., Bahçe M., Alikani M., Munné S. 2000. Chromosome abnormalities in 1255 cleavage-stage human embryos. Reprod. Biomed. Online 1, 17–26 (doi:10.1016/S1472-6483(10)61988-8) [DOI] [PubMed] [Google Scholar]
- Miller G. 2009. Neurodegeneration. Could they all be prion diseases? Science 326, 1337–1339 (doi:10.1126/science.326.5958.1337) [DOI] [PubMed] [Google Scholar]
- Mitalipova M. M., Rao R. R., Hoyer D. M., Johnson J. A., Meisner L. F., Jones K. L., Dalton S., Stice S. L. 2005. Preserving the genetic integrity of human embryonic stem cells. Nat. Biotechnol. 23, 19–20 (doi:10.1038/nbt0105-19) [DOI] [PubMed] [Google Scholar]
- Mosher J. T., Pemberton T. J., Harter K., Wang C., Buzbas E. O., Dvorak P., Simón C., Morrison S. J., Rosenberg N. A. 2010. Lack of population diversity in commonly used human embryonic stem-cell lines. N. Engl. J. Med. 362, 183–185 (doi:10.1056/NEJMc0910371) [DOI] [PubMed] [Google Scholar]
- Munné S., Tang Y. X., Grifo J., Rosenwaks Z., Cohen J. 1994. Sex determination of human embryos using the polymerase chain reaction and confirmation by fluorescence in situ hybridization. Fertil. Steril. 61, 111–117 [DOI] [PubMed] [Google Scholar]
- Nannya Y., et al. 2005. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 65, 6071–6079 (doi:10.1158/0008-5472.CAN-05-0465) [DOI] [PubMed] [Google Scholar]
- Närvä E., et al. 2010. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat. Biotechnol. 28, 371–377 (doi:10.1038/nbt.1615) [DOI] [PubMed] [Google Scholar]
- Oh S. K., et al. 2005. Derivation and characterization of new human embryonic stem cell lines: SNUhES1, SNUhES2, and SNUhES3. Stem Cells 23, 211–219 (doi:10.1634/stemcells.2004-0122) [DOI] [PubMed] [Google Scholar]
- Park J., Hwang S., Lee Y. S., Kim S. C., Lee D. 2007. SNP@Ethnos: a database of ethnically variant single-nucleotide polymorphisms. Nucleic Acids Res. 35, D711–D715 (doi:10.1093/nar/gkl962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson W. P. 2000. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays 22, 452–459 (doi:10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K) [DOI] [PubMed] [Google Scholar]
- Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., Smigielski E. M., Sirotkin K. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (doi:10.1093/nar/29.1.308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skottman H., Hovatta O. 2006. Culture conditions for human embryonic stem cells. Reproduction 132, 691–698 (doi:10.1530/rep.1.01079) [DOI] [PubMed] [Google Scholar]
- Song H., Chung S. K., Xu Y. 2010. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell 6, 80–89 (doi:10.1016/j.stem.2009.11.016) [DOI] [PubMed] [Google Scholar]
- Spits C., Mateizel I., Geens M., Mertzanidou A., Staessen C., Vandeskelde Y., Van der Elst J., Liebaers I., Sermon K. 2008. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat. Biotechnol. 26, 1361–1363 (doi:10.1038/nbt.1510) [DOI] [PubMed] [Google Scholar]
- St John J. C., Ramalho-Santos J., Gray H. L., Petrosko P., Rawe V. Y., Navara C. S., Simerly C. R., Schatten G. P. 2005. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells 7, 141–153 (doi:10.1089/clo.2005.7.141) [DOI] [PubMed] [Google Scholar]
- Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (doi:10.1126/science.282.5391.1145) [DOI] [PubMed] [Google Scholar]
- Tlsty T. D. 1998. Cell-adhesion-dependent influences on genomic instability and carcinogenesis. Curr. Opin. Cell Biol. 10, 647–653 (doi:10.1016/S0955-0674(98)80041-0) [DOI] [PubMed] [Google Scholar]
- Unger C., Skottman H., Blomberg P., Dilber M. S., Hovatta O. 2008. Good manufacturing practice and clinical-grade human embryonic stem cell lines. Hum. Mol. Genet. 17, R48–R53 (doi:10.1093/hmg/ddn079) [DOI] [PubMed] [Google Scholar]
- Van Hoof D., Braam S. R., Dormeyer W., Ward-van Oostwaard D., Heck A. J., Krijgsveld J., Mummery C. L. 2008. Feeder-free monolayer cultures of human embryonic stem cells express an epithelial plasma membrane protein profile. Stem Cells 26, 2777–2781 (doi:10.1634/stemcells.2008-0365) [DOI] [PubMed] [Google Scholar]
- Venter J. C., et al. 2001. The sequence of the human genome. Science 291, 1304–1351 (doi:10.1126/science.1058040) [DOI] [PubMed] [Google Scholar]
- Voullaire L., Collins V., Callaghan T., McBain J., Williamson R., Wilton L. 2007. High incidence of complex chromosome abnormality in cleavage embryos from patients with repeated implantation failure. Fertil. Steril. 87, 1053–1058 (doi:10.1016/j.fertnstert.2006.11.043) [DOI] [PubMed] [Google Scholar]
- Wells D., Bermudez M. G., Steuerwald N., Malter H. E., Thornhill A. R., Cohen J. 2005. Association of abnormal morphology and altered gene expression in human preimplantation embryos. Fertil. Steril. 84, 343–355 (doi:10.1016/j.fertnstert.2005.01.143) [DOI] [PubMed] [Google Scholar]
- Werbowetski-Ogilvie T. E., et al. 2009. Characterization of human embryonic stem cells with features of neoplastic progression. Nat. Biotechnol. 27, 91–97 (doi:10.1038/nbt.1516) [DOI] [PubMed] [Google Scholar]
- Wu H., et al. 2008. Copy number variant analysis of human embryonic stem cells. Stem Cells 26, 1484–1489 (doi:10.1634/stemcells.2007-0993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Inokuma M. S., Denham J., Golds K., Kundu P., Gold J. D., Carpenter M. K. 2001. Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 19, 971–974 (doi:10.1038/nbt1001-971) [DOI] [PubMed] [Google Scholar]
- Ylstra B., van den Ijssel P., Carvalho B., Brakenhoff R. H., Meijer G. A. 2006. BAC to the future! or oligonucleotides: a perspective for micro array comparative genomic hybridization (array CGH). Nucleic Acids Res. 34, 445–450 (doi:10.1093/nar/gkj456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis J. J. 1976. High resolution of human chromosomes. Science 191, 1268–1270 (doi:10.1126/science.1257746) [DOI] [PubMed] [Google Scholar]
- Zhao T., Xu Y. 2010. p53 and stem cells: new developments and new concerns. Trends Cell Biol. 20, 170–175 (doi:10.1016/j.tcb.2009.12.004) [DOI] [PubMed] [Google Scholar]
- Zhao X., et al. 2004. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 64, 3060–3071 (doi:10.1158/0008-5472.CAN-03-3308) [DOI] [PubMed] [Google Scholar]