Abstract
Long term survival of the pathogen Mycobacterium tuberculosis in humans is linked to the immunomodulatory potential of its complex cell wall glycolipids, which include the phosphatidylinositol mannoside (PIM) series as well as the related lipomannan and lipoarabinomannan glycoconjugates. PIM biosynthesis is initiated by a set of cytosolic α-mannosyltransferases, catalyzing glycosyl transfer from the activated saccharide donor GDP-α-d-mannopyranose to the acceptor phosphatidyl-myo-inositol (PI) in an ordered and regio-specific fashion. Herein, we report the crystal structure of mannosyltransferase Corynebacterium glutamicum PimB′ in complex with nucleotide to a resolution of 2.0 Å. PimB′ attaches mannosyl selectively to the 6-OH of the inositol moiety of PI. Two crystal forms and GDP- versus GDP-α-d-mannopyranose-bound complexes reveal flexibility of the nucleotide conformation as well as of the structural framework of the active site. Structural comparison, docking of the saccharide acceptor, and site-directed mutagenesis pin regio-selectivity to a conserved Asp residue in the N-terminal domain that forces presentation of the correct inositol hydroxyl to the saccharide donor.
Keywords: Cell Wall, Glycoconjugate, Membrane Lipids, Protein Structure, X-ray Crystallography, Corynebacterium glutamicum, Mycobacterium tuberculosis, Glycosyltransferase
Introduction
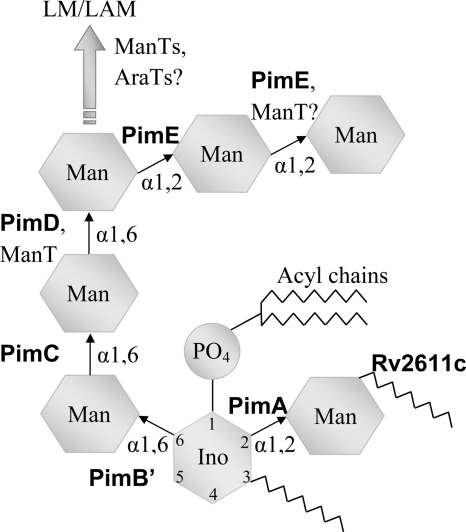
The cell envelope of Mycobacterium tuberculosis, the infectious agent causing tuberculosis, contains a variety of glycolipids that play a central role in subverting the host's immune response and thus help establish a long lasting latent infection, a hallmark of the pathophysiology of tuberculosis (1, 2). Phosphatidylinositol mannosides (PIMs)4 represent a series of glycolipids that comprise a phosphatidyl-myo-inositol (PI) core, an acylated mannosyl group attached to the 2-hydroxyl of inositol, and a mannosyl-oligosaccharide of variable length attached to the inositol 6-hydroxyl (Fig. 1) (3). A precursor of the more complex lipomannan (LM) and lipoarabinomannan (LAM) glycolipids, PIMs have been shown to influence both innate (4, 5) and adaptive immunity of the host (6–9).
FIGURE 1.
Schematic diagram of PIMs and pathway of PIM synthesis. ManT and AraT, generic mannosyl- and arabinosyltransferase enzyme, respectively. LM, lipomannan; LAM, lipoarabinomannan.
PIM biosynthesis begins by consecutive transfer of two mannosyl units from activated sugar-nucleotide (GDP-Man) to PI, catalyzed by cytoplasmic α-mannosyltransferases (Fig. 1). Attachment of the first mannosyl residue to the 2-hydroxyl of the inositol ring, resulting in PIM1, is catalyzed by PimA (M. tuberculosis Rv2610c, Mycobacterium smegmatis MSMEG_2935) (10, 11), followed by acylation of the 2-mannose (by M. tuberculosis Rv2611c) (12) to yield monoacylated PIM1 (Ac1PIM1). The second mannosyl residue is attached at the 6-hydroxyl, to yield Ac1PIM2, a reaction catalyzed by PimB′ (M. tuberculosis Rv2188c, M. smegmatis MSMEG_4253, Corynebacterium glutamicum NCgl2106) (13–15). The designation PimB had originally been assigned to open reading frame Rv0557, also encoding an α-mannosyltransferase. Subsequently, Rv0557 was found to be dispensable for synthesis of Ac1PIM2, and the corresponding protein has since been renamed MgtA (15–17). For consistency with the recent literature, we retain the designation PimB′ for Rv2188c (and its orthologs in M. smegmatis and C. glutamicum) (14, 15, 18).
Prolonged incubation of Ac1PIM2 with PimB′ or PimA does not lead to further extension of Ac1PIM2 (14). Instead, extending the mannosyl chain at the 6-OH requires a distinct set of mannosyltransferases. For instance, bioinformatic analysis of the genome of M. tuberculosis CDC1551 led to the identification of PimC, which catalyzes synthesis of the trimannoside Ac3PIM3 (19). Still, the pimC deletion in Mycobacterium bovis bacille Calmette-Guérin does not interrupt formation of higher PIMs, LM or LAM, for which PIMn (where n = 1–3) is considered a precursor, whereas genes orthologous to pimC were found in only 22% of clinical isolates. Thus, compensatory synthetic pathways must exist. Higher order PIM, LM, and LAM depend on glycosyl transfer from the lipid-saccharide donor C30/C50-P-Man and membrane-embedded glycosyltransferases, including PimE (20), with evidence for pathway bifurcation at Ac2PIM4 (21, 22).
The three-dimensional structures of soluble glycosyltransferases display only two fundamental fold topologies, termed GT-A and GT-B (23), contrasting with the diversity of protein folds among glycoside hydrolase enzymes (24). According to the sequence-based classification of carbohydrate-active enzymes, PimB′ (Rv2188c) belongs to glycosyltransferase family 4 (GT4; see the CAZy database) (25)). Encompassing a diverse range of enzymatic activities and substrate specificities, GT4 family enzymes assume the GT-B fold, with known structures for about half a dozen family members (26–30). The GT-B topology is characterized by two Rossmann fold-like domains, where donor and acceptor substrates bind in the central cleft between the two domains. In GT4 family transferases, the C-terminal domain provides the majority of contacts for the nucleoside-diphosphate-saccharide donor substrate, whereas the diverse acceptor-substrates bind to the more variable N-terminal domain. A hinge region allows for conformational flexibility between the two domains, which can be dramatic, as is illustrated by the structures of ligand-free and substrate-bound C. glutamicum MshA (Protein Data Bank entries 3c48 and 3c4v (29)). Between these two states, the relative orientation of the domains changes by a 97° rotational movement. Similar if less dramatic examples of interdomain flexibility have been observed in structures of several other GT4 family members.
Recently determined structures of GT4 family enzymes include that of M. smegmatis PimA, the enzyme catalyzing transfer of the first mannosyl group to PI (27). This structure shed light on the mode of nucleotide binding and suggested a model for recognition of the acceptor substrate. Although PimA and PimB′ utilize the same donor substrate, their acceptor specificity is distinct. PimB′ appears unable to mannosylate phosphatidyl-myo-inositol (or myo-inositol-1-phosphate) on the 2-hydroxyl, whereas PimA does not mannosylate PI or PIM1 on the 6-hydroxyl.
In order to clarify structural differences between PimA and PimB′ that could explain regio-selectivity, we undertook the structure determination of PimB′. Attempts to generate recombinant protein of M. tuberculosis PimB′ (Rv2188c) were unsuccessful, but overexpression of the C. glutamicum ortholog (NCgl2106) in Escherichia coli yielded soluble protein that crystallized when incubated with the donor substrate. Herein, we report the crystal structure of C. glutamicum PimB′ in complex with nucleotide to a resolution of 2.0 Å, revealing flexibility of both the nucleotide conformation and the structural framework of the active site. The results of our site-directed mutational analysis are consistent with a substrate-mediated SNi (internal return) reaction mechanism and indicate that a conserved aspartic acid in the acceptor-binding domain is critical in determining regio-selectivity of mannosyl transfer.
EXPERIMENTAL PROCEDURES
Recombinant Protein Production
Cloning of C. glutamicum PimB′ has been described previously (15). Liquid cultures of E. coli BL21 (DE3), harboring pET16b-pimB′, were grown at 30 °C in LB broth medium (Difco), supplemented with ampicillin (100 μg/ml). Expression of pimB′ was induced at A600 0.4–0.6 by adding 0.5 mm isopropyl-β-d-thiogalactoside. Cultures were incubated for a further 4 h before being harvested (4 °C, 4000 × g), washed with 0.85% saline, and stored at −20 °C. For the expression of SeMet-labeled PimB′, liquid cultures were grown to A600 0.4 and then pelleted and resuspended in 1 liter of minimal medium twice (50 mm Na2HPO4, 20 mm KH2PO4, 10 mm NaCl, 20 mm NH4Cl, 20 mm MgSO4, 0.1 mm CaCl2, and 0.4% (w/v) glucose). Cells were grown at 37 °C to A600 0.5, and then 100 mg of l-lysine, l-phenylalanine, and l-threonine; 50 mg of isoleucine, leucine, and valine; and 60 mg of l-selenomethionine were added. After 30 min at 37 °C, expression of pimB′ was induced, adding 0.5 mm isopropyl-β-d-thiogalactoside, followed by incubation at 25 °C for 24 h. Cells were harvested and treated as before.
Purification and Crystallization
Cell pellets were thawed on ice and resuspended in 30 ml of 50 mm NaH2PO4, pH 8, 300 mm NaCl, and 10 mm imidazole (Buffer A), supplemented with an EDTA-free protease mixture tablet (Roche Applied Science). The suspension was sonicated (Sonicator Ultrasonic Liquid Processor XL, Misonix Inc.) on ice for a total time of 10 min, using 20-s pulses and 40-s cooling periods. The lysate was centrifuged (27,000 × g, 30 min, 4 °C), and the supernatant was passed through a Buffer A-equilibrated Ni2+-charged His-Trap column (1 ml; GE Healthcare). The column was subsequently washed with 50 ml of Buffer A with 20 mm imidazole, and the protein was eluted with a 50–300 mm step gradient of imidazole. Fractions containing protein were dialyzed against 20 mm Tris-HCl, pH 7.5, 10 mm NaCl, 10% glycerol, and 1 mm DTT. After dialysis, proteins were concentrated by ultrafiltration to 8 mg/ml using a 10 kDa cut-off (Centriprep, Millipore).
Prior to setting up crystallization trials, N-terminally tagged PimB′ was incubated overnight with 10 mm GDP-Man. Commercial sparse matrix screens (Molecular Dimensions) were used to identify crystallization conditions in hanging drop vapor diffusion experiments. Thin plate crystals formed at 6–26% PEG 3350, 0.1 m BisTris, pH 5.5, 0.1 m lithium sulfate, and 0.1 m glycine. Crystals were cryoprotected by adding reservoir solution supplemented with 8% (w/v) sucrose and 8% (v/v) glycerol to the crystallization drop with subsequent flash freezing in liquid nitrogen. Crystals of N-terminally tagged PimB′ were in space group P1, diffracting to 3.5 Å on the in-house x-ray source (Rigaku MicroMax 007HF, Saturn 944 CCD) and to 2.2 Å at the synchrotron.
Structure Determination of Triclinic Crystal Form
Single wavelength data of native C. glutamicum PimB′ were recorded on beamline ID29 at the European Synchrotron Radiation Facility (ESRF) (Grenoble, France) (Table 1) at 10% transmission. Analysis of the Matthews volume suggested the presence of two molecules of PimB′ per asymmetric unit (VM = 2.28 Da/Å3) with a predicted solvent fraction of 47%. Initial attempts to phase by molecular replacement resulted in a statistically significant solution in terms of the PHASER Z-score (Z = 9.0, resolution range 10–3 Å) (31) but failed to yield a useful electron density map. Because the thin plate crystals did not tolerate soaking in heavy metal salts, we generated SeMet-derivatized crystals of PimB′ and recorded data at the Se-K edge (Table 1). Significant radiation damage of the cryogenically cooled crystals limited data acquisition to the peak wavelength and, due to the triclinic symmetry, resulted in data of low multiplicity (Table 1). In order to optimize the anomalous signal and completeness of Bijvoet pairs, an inverse beam protocol was used, recording data in sets of 20 successive frames (0.5° oscillation) at φ and φ + 180°, respectively. Data were reduced using XDS and XSCALE (32). Only one of several SeMet single-wavelength anomalous dispersion data sets (SeMet 1 in Table 1) recorded from a series of crystals provided a sufficiently strong anomalous signal to extract the selenium positions using SHELXD (33). Patterson self-rotation maps indicated a non-crystallographic (NCS) 2-fold rotation axis parallel to the b-axis, consistent with the assumption of two molecules per asymmetric unit. Therefore, selenium positions obtained in runs of SHELXD were analyzed with PROFESSS (34) to derive the NCS operator and to verify that this operator was consistent with the self-rotation function. An initial set of 10 selenium positions (five per molecule) could be identified this way. Single-wavelength anomalous dispersion phasing in SHARP (35) allowed us to establish an additional set of eight positions through difference Fourier analysis. Thus, 18 of 20 possible selenium positions were obtained but were not sufficient to obtain an interpretable map, either in single-wavelength anomalous dispersion phasing using the best SeMet data or in phasing by multiple isomorphous replacement using the best SeMet together with the native data.
TABLE 1.
Data acquisition and structure refinement statistics
| Data acquisition | PimB′ + GDP-Man | SeMet 1 | SeMet 2 | SeMet 3 | SeMet 4 | PimB′ + Br-GDP-Man | PimB′ + GDP-Man | PimB′ + GDP |
|---|---|---|---|---|---|---|---|---|
| Beamline | ESRF ID29 | Diamond I04 | Diamond I04 | ESRF ID23.1 | ESRF ID29 | Diamond I02 | Diamond I04 | ESRF ID14.2 |
| Wavelength (Å) | 0.9762 | 0.9805 | 0.9805 | 0.97905 | 0.97888 | 0.9201 | 0.9763 | 0.933 |
| Space group | P1 | P1 | P1 | P1 | P1 | P1 | P212121 | P212121 |
| Cell parameters a, b, c (Å) | 44.1, 50.0, 85.3 | 44.1, 50.1, 85.3 | 44.1, 50.0, 85.3 | 43.9, 49.9, 85.4 | 44.0, 50.0, 85.7 | 43.9, 49.9, 85.7 | 44.8, 87.0, 106.9 | 44.8, 86.3, 106.4 |
| Cell parameters α, β, γ | 92.1, 92.7, 89.9 | 92.2, 92.6, 90.1 | 92.0, 92.8, 90.0 | 91.9, 92.4, 90.1 | 92.2, 92.6, 90.0 | 92.1, 92.7, 89.9 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 44.0-2.2 | 29.96-2.90 | 29.91-2.90 | 25.38-2.90 | 29.97-2.80 | 29.91-2.90 | 37.29-2.0 | 45.37-2.23 |
| High resolution shell (Å) | 2.32-2.20 | 3.06-2.90 | 3.06-2.90 | 3.06-2.90 | 2.95-2.80 | 3.06-2.90 | 2.11-2.00 | 2.35-2.23 |
| Rmerge (%)a | 7.3 (23.8) | 4.9 (11.1) | 7.1 (15.5) | 6.7 (11.3) | 8.5 (18.1) | 11.6 (26.2) | 9.6 (38) | 9.3 (38.2) |
| Total no. of observationsa | 103582 (12352) | 40261 (5462) | 46289 (6804) | 44487 (6484) | 52162 (7446) | 46058 (6569) | 208274 (29919) | 64840 (4871) |
| Unique reflectionsa | 35191 (4400) | 13101 (1951) | 13931 (2092) | 13532 (2027) | 17456 (2507) | 15023 (2197) | 28997 (4147) | 19982 (2147) |
| 〈I/σ(I)〉a | 11.6 (5.9) | 18.1 (7.1) | 11.6 (5.3) | 12.5 (7.8) | 9 (5.2) | 7.2 (3.2) | 18.3 (5.7) | 9.9 (2.3) |
| Completeness (%)a | 95.1 (81.2) | 81 (83.1) | 86.4 (89.3) | 84 (86.4) | 97.4 (95.8) | 93.4 (93.4) | 100 (99.8) | 95.8 (72.5) |
| Multiplicitya | 2.9 (2.8) | 3.1 (2.8) | 3.3 (3.3) | 3.3 (3.2) | 3 (3) | 3.1 (3.0) | 7.2 (7.2) | 3.2 (2.3) |
| Anomalous completeness (%)a | 70.5 (70.0) | 84.5 (87.1) | 77.3 (80.3) | 92.5 (90.1) | 82.4 (82.7) | |||
| Anomalous multiplicitya | 1.7 (1.6) | 1.7 (1.7) | 1.7 (1.7) | 1.5 (1.5) | 1.7 (1.6) | |||
| Δano correlation between half-setsa | 0.483 (0.174) | 0.272 (0.138) | 0.265 (0.132) | 0.246 (0.198) | 0.101 (0.073) |
| Refinement | PimB′ + GDP-Man | PimB′ + GDP-Man (orth) | PimB′ + GDP (orth) |
|---|---|---|---|
| Space group | P1 | P212121 | P212121 |
| Resolution range (Å) | 44.0–2.2 | 37.3–2.0 | 45.4–2.4 |
| Unique reflections | 33243 | 27519 | 15991 |
| Rcryst (%) | 18.9 | 20.3 | 19 |
| Rfree (%)b | 22.1 | 23.2 | 23.6 |
| No. of non-hydrogen atoms | |||
| Total | 6235 | 3068 | 3019 |
| Protein | 5829 | 2829 | 2851 |
| Ligand | 56 | 56 | 28 |
| Solvent | 350 | 183 | 140 |
| r.m.s. deviation bonds (Å)c | 0.008 | 0.007 | 0.007 |
| r.m.s. deviation angles (degrees)c | 1.15 | 1.02 | 1.09 |
| B-factor (Å2) | |||
| Wilson | 17.3 | 15.9 | 28 |
| Average | 13.1 | 14.2 | 20.4 |
| Protein | 12.2 | 13.8 | 20.1 |
| Ligand | 31.9 | 20.6 | 38.2 |
| Solvent | 17.5 | 19.3 | 24.7 |
| r.m.s. deviation B-factor | |||
| Between bonded main chain atoms | 0.42 | 0.26 | 0.34 |
| Bonded side chain atoms | 1.17 | 0.65 | 0.55 |
| Ramachandran plotd | |||
| Favored (%) | 98.7 | 98.1 | 98.0 |
| Outliers (no.) | 0 | 0 | 0 |
Next, we co-crystallized PimB′ (N-terminally tagged) with a brominated derivative of GDP-Man, the synthesis of which has been described (36). Combining the PimB′ native data with this brominated derivative and the best four SeMet data sets led to a density map with recognizable secondary structure features (supplemental Fig. 2SA), albeit still too poor to build. 2-fold NCS averaging and phase extension to nominally 2.2 Å distinctly improved the map (supplemental Fig. 2SB). In order to support map interpretation, we reconciled the previously established molecular replacement solution with the experimentally determined set of selenium positions. This was aided by generating a homology model of the C-terminal domain of PimB′ (using MODELLER (37)), which reproduced a subset of distance vectors between selenium positions. Superimposing the homology model onto the experimental map aided interpretation of poorly defined areas, greatly accelerating model building. Iterative rounds of model building and minimization in REFMAC5 (38) very quickly led to a high quality 2Fo − Fc map (supplemental Fig. 2SC) that revealed previously undefined parts of the structure. The final model describes residues 4–381 of the PimB′ (381 residues), with excellent stereochemistry (Table 1).
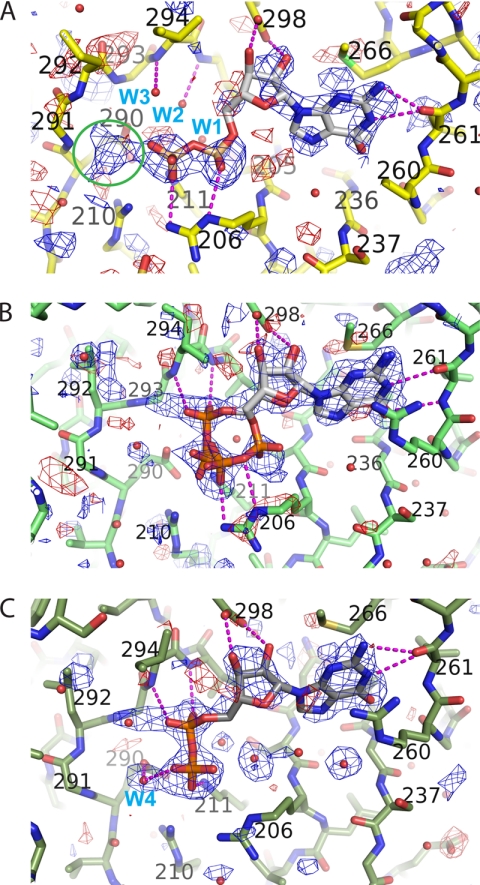
In both molecules of the triclinic crystals, the active site contains weak density for GDP-Man. Therefore, the ligand was omitted from the model until the structure refinement had converged to Rcryst/Rfree values of 19.1%/22.4%. Adding GDP to this late stage model, at full occupancy, reduced Rfree marginally, giving rise to negative difference density. Systematic variation of the occupancy of GDP between 0.5 and 1 resulted in a flat difference map around the ligand for 0.75. At this occupancy, the B-factors of the ligand atoms refined to an average of 31 Å2, compared with a protein average of 12.5 Å2 and an overall Wilson B-factor of 19.3 Å2. Although density for the mannose moiety is visible (see Fig. 4A), it is insufficient to fit the saccharide unequivocally.
FIGURE 4.
Different modes of nucleotide binding in the active site of PimB′. A, σA-weighted Fo − Fc density map (contour levels 2.2 σ (blue) and −2.2 σ (red)) of triclinic PimB′. The green circle marks the position of the density blob corresponding to the GDP-linked mannose. B and C, σA-weighted Fo − Fc density maps (+2.8 σ (blue), −2.8 σ (red)) of GDP-Man (B) and GDP-bound (C) complexes of orthorhombic PimB′. Selected water molecules are labeled in blue, and dashed lines in magenta indicate a subset of relevant non-covalent interactions. All maps were calculated with Fc amplitudes and phases of the ligand-free model.
Structure Determination of the Orthorhombic Crystal Form
C-terminally tagged PimB′ (8 mg/ml) was incubated overnight with either GDP or GDP-Man (20 mm) prior to setting up vapor diffusion experiments. Crystals of sufficient size appeared over reservoirs containing 0.1 m dl-malic acid, pH 6.1, 13% PEG 3350 (GDP-Man complex) or 0.1 m sodium citrate, pH 5.1, 22% PEG 3350 (GDP complex). Crystals were cryoprotected in mother liquor supplemented with 8% (w/v) sucrose and 8% glycerol and frozen in liquid nitrogen. The structure of the orthorhombic crystal form of PimB′ was solved by molecular replacement, using a monomer of the refined “triclinic” structure as search model in PHASER (31). The structural models were rebuilt in COOT and refined using REFMAC5 (38). Figs. 2–5 were prepared using PyMOL (DeLano Scientific LLC).
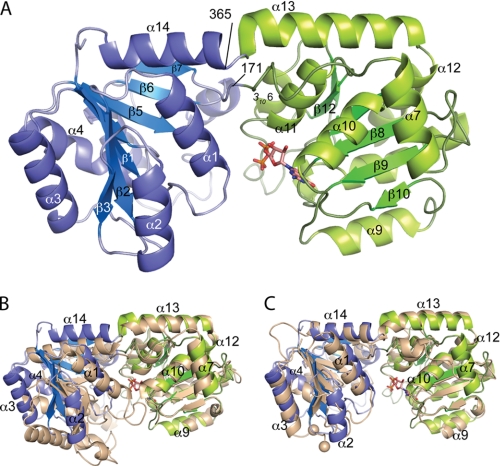
FIGURE 2.
Ribbon diagram of C. glutamicum PimB′ in the triclinic crystal form and comparison with PimA and MshA. A, N-terminal domain (blue) and C-terminal domain (green) are shown together with a stick model of the bound nucleotide. B, comparison with C. glutamicum MshA (Protein Data Bank entry 3c4v; beige) after superposition with respect to secondary structure elements in the C-terminal domain. C, comparison with the structure of PimA (beige) following domain-wise superposition onto PimB′. Beige spheres indicate the ends of the disordered “IP-binding” loop of PimA.
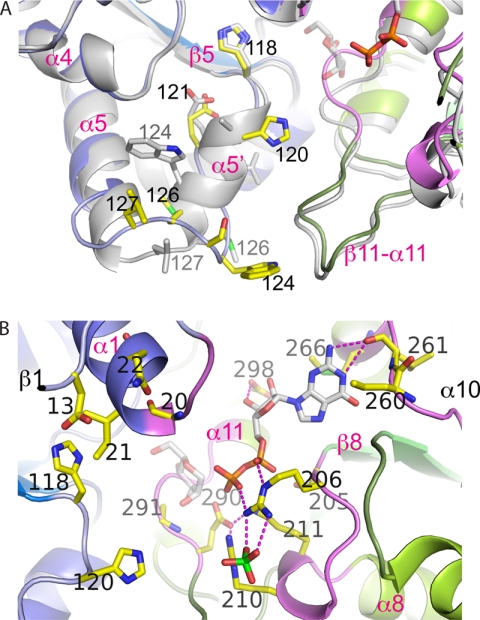
FIGURE 3.
Structural flexibility in the β5-α5 loop and overview of the active site of PimB′. A, comparison of conformation between the triclinic (green/blue) and orthorhombic crystal (gray) forms of PimB′. Helix α5′ is only present in the orthorhombic form. B, close-up view of the active site cleft of triclinic PimB′ in the open configuration with ribbons in blue and green indicating the N- and C-terminal domains, respectively. Loop regions involved in nucleotide binding are colored in magenta. Residues in contact with nucleotide or that were probed by point mutagenesis are shown in yellow sticks. The nucleotide shown reflects the “PimA-like” conformation of the pyrophosphate. The mannose moiety is shown in transparent sticks, modeled according to the conformation seen in OtsA.
FIGURE 5.
Model of the closed form of PimB′ with docked acceptor. A, ribbon diagram of the closed configuration of PimB′ superimposed with the molecular surface of the open structure of GDP-Man-bound PimB′ (orthorhombic crystal form). The dashed line (black) indicates the approximate position of the membrane surface. B, close-up view of the docked ternary complex. PIM1 (cyan bonds) is shown without the acyl chains and the glycerol backbone. Side chains probed by mutagenesis are show in blue (N-terminal domain) and green (C-terminal domain).
Preparation of Polar Lipids and Ac1PIM1
Polar lipids containing Ac1PIM1 were extracted from C. glutamicum-ΔpimB′ΔmgtA as described previously (15, 39). The lipid extract was examined by two-dimensional TLC on aluminum-backed plates of Silica Gel 60 F254 (Merck 5554), using CHCl3/CH3OH/H2O (60:30:6, v/v/v) in the first direction and CHCl3/CH3COOH/CH3OH/H2O (40:25:3:6, v/v/v/v) in the second direction. The glycolipids were visualized by spraying plates with either α-naphthol/sulfuric acid, Dittmer and Lester reagent, or 5% ethanolic molybdophosphoric acid followed by gentle charring of plates. Glycolipids were further separated into individual PIMs and other glycolipid species by preparative TLC on 10 cm × 20-cm plastic-backed TLC plates of Silica Gel 60 F254 (Merck 5554), run in chloroform/methanol/water (60:30:6, v/v/v). The plates were then sprayed with 0.01% 1,6-diphenyl-1,3,5-hexatriene dissolved in petroleum ether/acetone (9:1, v/v), and the glycolipids were visualized under UV light. Following detection, the plates were redeveloped in toluene to remove diphenyl-1,3,5-hexatriene, and the corresponding glycolipid bands were scraped from the plates and extracted from the silica gel using chloroform/methanol (2:1, v/v). Samples were analyzed by MALDI-TOF MS as described previously (17).
Activity Assay
PimB′ was assayed for mannosyltransferase activity using a method adapted from Ref. 40. 20 μg of PimB′ was incubated with either purified Ac1PIM1 (4–200 μg) or polar lipids (100 μg) extracted from C. glutamicum-ΔpimB′ΔmgtA, in the presence of 0.5 μCi of GDP-[14C]mannose (GE Healthcare), 0.125 mm ATP, 1 mm NADP, 10 mm CaCl2, 0.1% (v/v) IGEPAL (Sigma), and 20 mm Tris-HCl, pH 7.5, in a total volume of 100 μl. The reaction was incubated at 37 °C for 30 min and quenched with the addition of CHCl3/MeOH/H2O (10:10:3). Unincorporated GDP-[14C]mannose was removed by the addition of 2.625 ml of CHCl3 and 1.125 ml of H2O, followed by centrifugation to separate the organic and aqueous layers. The lower organic layer was washed three times with 3 ml of CHCl3/MeOH/H2O (3:47:48) and dried. Incorporation of GDP-[14C]mannose in the acceptor was measured using a scintillation counter, and samples were analyzed by TLC run in CHCl3/MeOH/H2O/NH4OH (65:25:3.6:0.5).
Trp Fluorescence Binding Assay
Binding of GDP and GDP-Man was measured using intrinsic tryptophan fluorescence. The sample was excited at a wavelength of 295 nm, and the resulting fluorescence emission was measured between 300 and 400 nm (scan speed 100 nm/min) using a Photon Technology International fluorescence spectrometer. Spectra were recorded for each addition of GDP-Man or GDP to 700 μl of a 2 μm solution of PimB′. The experiment was repeated in triplicate, and fluorescence intensity at 330 nm was plotted against ligand concentration and fitted to a one-site saturation binding model using SIGMAPLOT (Systat). In order to compensate for inner filtering of the ligand, we monitored fluorescence of albumin at a range of GDP concentrations and derived a concentration-dependent correction to the Trp fluorescence signal.
Docking of PIM1
A molecular model of PIM1 was manually docked into the active site of PimB′ (closed configuration) guided by the following considerations: (i) no steric clashes must occur, and no adjustments to the protein structure should be necessary; (ii) the 6-hydroxyl of inositol should be presented to C1 of the donor saccharide while minimizing the oxygen-carbon distance; (iii) if possible, inositol hydroxyls should form H-bond contacts with nearby H-bond acceptors; (iv) while bound to the enzyme, the acyl chains attached to the glycerol backbone of the acceptor should be free to insert into the cell membrane. The validity of the resulting model of the ternary complex was tested by mutagenesis.
RESULTS
Structure Determination
Crystals of C. glutamicum PimB′ were grown in two crystal forms with triclinic and orthorhombic lattices, depending on whether the recombinant protein carried a His6 affinity tag at the N or C terminus, respectively. Incubation of N-terminally tagged PimB′ with the native donor substrate guanosine-5′-diphosphate-α-d-mannopyranose (GDP-Man; 10 mm) produced triclinic crystals, based on which the structure was determined (see “Experimental Procedures”) (supplemental Fig. 2S). The refined “triclinic” model (2.2 Å resolution; Fig. 2A and Table 1) comprises two molecules of PimB′ (residues 4–381) in the asymmetric unit, missing only the first three residues of C. glutamicum PimB′. The two copies were refined applying tight NCS restraints (root mean square (r.m.s.) deviation between 378 Cα pairs of 0.03 Å, estimated coordinate error 0.37 Å; Table 1). Subsequent to the structure determination, an orthorhombic crystal form was obtained and refined to a resolution of 2.0 Å. The lattices of the orthorhombic and triclinic crystal forms are similar (triclinic, at = 44 Å, bt = 50 Å, ct = 85 Å; α ≈ β ≈ γ ≈ 90°; orthorhombic, ao = at, bo = ct, co ≈ 2 bt) (Table 1) but show distinct packing interfaces. This is presumably due to the affinity tag, which is disordered in the orthorhombic lattice but partially ordered in the triclinic crystal form. Weak density for GDP-Man was observed in both copies of PimB′ in the “triclinic” model, suggesting partial occupancy. In contrast, the GDP- and GDP-Man-bound structures of the orthorhombic form show strong density for the nucleotide, although the mannose moiety is disordered (see below).
Overall Structure of PimB′
The structure of C. glutamicum PimB′ displays the canonical fold of GT-B glycosyltransferases, comprising two Rossmann fold-like domains (41) that are connected by a hinge region with the substrate binding sites located in the cleft between the domains (Fig. 2A). The N-terminal domain (residues 4–171 and 364–381), assumed to bind the acceptor substrate, is composed of a seven-stranded, parallel β-sheet (strands β1–β7), with connecting helices α1–α6. The C-terminal domain (residues 172–363) binds the nucleotide-sugar donor substrate and consists of a six-stranded, parallel β-sheet (strands β8–β13) with connecting helices α8–α11 and 3106. A conserved feature of GT4 family enzymes, and indeed of the GT-B fold, is the sequence of the two C-terminal helices α13 and α14 that span the length of the protein and belong to the C- and N-terminal domains, respectively. Domain reorientation is facilitated by a flexible hinge between the domains, consisting of the β7-α7 loop, with invariant Gly171 at its center, and the short linker separating helices α13 and α14, with highly conserved Trp365 clamped between the domains (Fig. 2A and supplemental Fig. 1S).
Differences in crystal packing between the orthorhombic and triclinic crystal forms resulted in a slightly different interdomain orientation (4° rotation) and an additional helix, α5′ (residues 120–125), in the active site cleft (Fig. 3A). In the domain-wise superposition, the triclinic and orthorhombic models match closely (r.m.s. deviation of 1.27 Å for the N-terminal domain, 0.39 Å for the C-terminal domain), but individual residues in the region of helix α5′ undergo considerable shifts of their position. For instance, the α-carbon of Trp124 is shifted by 7 Å, and its side chain switches from contacts with the C-terminal domain (Gly285 and Leu287 of the β11-α11 loop) to folding back onto the N-terminal domain in the orthorhombic form.
Structural Neighbors
We searched for structural homologs of PimB′ in the Protein Data Bank through distance matrix alignment (DALI (42)). The closest neighbor with respect to overall structure was glycosyltransferase MshA from C. glutamicum (Fig. 2B) (Protein Data Bank entry 3c48; Z = 23.7, r.m.s. deviation 1.6 Å, 180 aligned Cα pairs of 393 residues, sequence identity 31%) (29), followed by glycosyltransferases E. coli WaaG (Protein Data Bank entry 2iw1; Z = 21.8, r.m.s. deviation 2.3 Å, 180 of 370 Cα pairs, 25% identity) (26), and Bacillus anthracis Ba1558 (Protein Data Bank entry 2jjm; Z = 21.3, r.m.s. deviation 1.8 Å, 177 of 359 Cα pairs, 21% identity) (28). The fourth hit was the functionally related α-mannosyltransferase M. smegmatis PimA (Fig. 2C; Protein Data Bank entry 2gej, Z = 20.1, r.m.s. deviation 1.9 Å, 176 of 361 Cα pairs, 24% identity). Restricting the DALI search to the more variable N-terminal domain, which commonly binds the acceptor substrate, the closest neighbor was the N-terminal domain of B. anthracis Ba1558 (Protein Data Bank entry 2jjm, Z = 19.2, r.m.s. deviation 2.1 Å, 165 Cα, 18% identity), followed by those of MshA (Protein Data Bank entry 3c48, Z = 16, r.m.s. deviation 2.6 Å, 172 Cα, 10% identity) and PimA (Protein Data Bank entry 2gej, Z = 15.4, 161 Cα, 17% identity).
The structural comparison between PimB′ and PimA (Fig. 2C) is of particular interest because they catalyze subsequent steps in PIM biosynthesis utilizing identical saccharide donors (GDP-Man) yet showing distinct regio-selectivity for mannosylation of the inositol moiety of PI. The domain-wise superposition of PimB′ and PimA shows conserved cores for either domain (N-terminal domain, r.m.s. deviation 1.53 Å for 106 Cα pairs; C-terminal domain, r.m.s. deviation 1.19 Å for 147 Cα pairs). However, several structural differences are noteworthy. First, PimB′ lacks the extended β3-α3 loop, due to a 10-residue deletion (supplemental Fig. 1S). This loop, referred to as the “IP-binding” loop (10, 29), has been implicated in acceptor substrate binding in both PimA and MshA. Although fully ordered in PimB′, it is disordered in nucleotide-bound PimA (Fig. 2C). Second, the β11-α11 loop in PimB′ has a 6-residue insertion relative to PimA. This loop makes contacts with the structurally variable loop β5–α5 (containing helix α5′ in orthorhombic PimB′). Furthermore, PimB′ comprises additional helices in the N-terminal (α2) and C-terminal domains (α7; Fig. 2C), whereas helix α5 of PimB′ overlaps partially with helices α4 and α5 in PimA.
Active Site and Nucleotide Binding
We crystallized PimB′ in the presence of GDP or GDP-Man, which bind with affinities on the order of 20–40 μm in terms of Kd (Table 2). Density for the nucleotide is weak in the triclinic crystal form (10 mm GDP-Man present during crystallization; Fig. 4A) but is well defined for the orthorhombic crystals (20 mm GDP or GDP-Man; Fig. 4, B and C). The nucleotide binding site is situated in the cleft between N- and C-terminal domains of PimB′ (Fig. 3B), a conserved feature of GT-B glycosyltransferases. In all cases, binding of the nucleotide (GDP or GDP-Man) resulted in crystallization of an “open” configuration of PimB′, in marked contrast to the donor substrate-bound structures of PimA and MshA (supplemental Fig. 3S). In this configuration, the central cleft is open, and the nucleotide makes no contacts with residues in the N-terminal domain. A small but significant density blob in triclinic PimB′ indicates the presence of mannose (Fig. 4A), but the blob fails to define the conformation of the saccharide moiety unequivocally. Paradoxically, the mannose moiety is disordered in the orthorhombic crystals of GDP-Man-bound PimB′ (Fig. 4B), although the ligand density is much stronger overall. We attribute the absence of mannose density to the absence of stabilizing contacts in the open form of the enzyme. We found no indication of mannose release in the absence of acceptor substrate when incubating PimB′ with 14C-labeled GDP-Man over a period of several h.
TABLE 2.
Activity and saturation binding for PimB′ and selected point mutants
| GDP-Man Kd | GDP Kd | Activity | Residual Activity | |
|---|---|---|---|---|
| μm | μm | cpm | % | |
| WT | 19.0 ± 4.6 | 36.5 ± 6.9 | 5528 ± 979 | 100.0 |
| Blank | NAa | NA | 30 ± 22 | 0.5 |
| N12A | 20.4 ± 6.2 | 21.3 ± 5.5 | 1723 ± 77 | 31.2 |
| D13N | 30.1 ± 6.9 | 18.8 ± 5.1 | 83 ± 5 | 1.5 |
| D13Y | NA | NA | 16 ± 0 | 0.3 |
| D13A | 42.8 ± 11.9 | 61.3 ± 17.4 | 104 ± 5 | 1.9 |
| G20W | 21.9 ± 6.4 | 46.5 ± 40.2 | 176 ± 15 | 3.2 |
| I21S | 13.4 ± 7.0 | 26.5 ± 7.4 | 134 ± 22 | 2.4 |
| I21A | 56.0 ± 14.5 | 40.9 ± 8.6 | 110 ± 5 | 2.0 |
| Q22A | 60.3 ± 22.8 | 30.6 ± 6.4 | 788 ± 218 | 14.3 |
| H118S | 72.4 ± 27.5 | 19.6 ± 9.8 | 36 ± 3 | 0.7 |
| H120S | NA | NA | 4490 ± 524 | 81.2 |
| G123P | 29.3 ± 5.9 | 42.3 ± 9.3 | 4784 ± 298 | 86.6 |
| S205G | 18.7 ± 4.6 | 33.6 ± 6.9 | 6609 ± 1207 | 119.6 |
| R206S | 42.7 ± 9.4 | 72.4 ± 17.2 | 311 ± 47 | 5.6 |
| R210S | 40.2 ± 10.7 | 44.1 ± 28.8 | 481 ± 58 | 8.7 |
| K211Q | 30.5 ± 17.6 | 40.3 ± 14.6 | 193 ± 25 | 3.5 |
| E290Q | 35.7 ± 18.9 | 41.7 ± 21.5 | 252 ± 30 | 4.6 |
| E290D | 55.0 ± 17.7 | 322.7 ± 518.7 | 162 ± 14 | 2.9 |
| E290N | NBb | 59 ± 16 | 1.1 | |
| G291S | 68.7 ± 35.5 | 74.2 ± 37.2 | 292 ± 241 | 5.3 |
a NA, binding not assayed.
b NB, no binding detected.
With few exceptions, residues in contact with the nucleotide reside in the following structural elements: the β8-α8 loop, the β10-α10 loop, and the β11-α11 loop (Fig. 3B). The purine base forms hydrogen bonds with the amide nitrogen and carbonyl groups of Leu261 (β10-α10 loop) but no specificity-determining contacts with side chain atoms. The side chains of Val235 (C-terminal end of β9), Arg260, and Met266 (β10-α10 loop) contact the base by way of apolar and van der Waals contacts. Interestingly, in the (low occupancy) “triclinic” structure, Arg260 is only partially ordered. The C2′ and C3′ hydroxyls of the ribose make conserved H-bond interactions with the side chain carboxyl of Glu298 (helix α11). The pyrophosphate group is set against a surface of positive electrostatic potential of residues Arg206, Arg210, and Lys211 (β8-α8 loop) and the “oxyanion hole”-like surface of four consecutive amide nitrogen groups (residues 291–294) in the β11-α11 loop. The carboxylate group of Glu290, a highly conserved residue in the β11-α11 loop, is held in a fixed orientation through ionic bonds with Lys211 and Arg210, neutralizing its negative charge that would otherwise repel the close-by β-phosphate (distance ∼3.8 Å).
It was surprising to find the pyrophosphate in two distinct conformations that differ with respect to the position of the α-phosphate, whereas the position of the β-phosphate is more or less fixed, stabilized by ionic contacts with the guanido group of Arg206 and the terminal ammonium group Lys211 (Figs. 3B and 4). Recording diffraction data of three independent GDP-bound complexes, the pyrophosphate of GDP consistently displayed a conformation that coincides with that seen in the UDP-bound complex of trehalose-6-phosphate synthase (E. coli OtsA; Protein Data Bank entries 1uqu and 2wtx (43–45)). In this “OtsA-like” conformation, the α-phosphate is positioned close to the main chain amide nitrogen groups of Ile294 and Val295 (N-terminal of helix α11), forming weak H-bonds (3.3–3.5 Å) (Fig. 4C). In contrast, orthorhombic crystals of PimB′ in complex with GDP-Man show a superposition of the “OtsA-like” conformer and a second conformation that resembles the nucleotide conformation in GDP-Man-bound PimA (Fig. 4B) (Protein Data Bank entries 2gek and 2gej) (27). In the PimA-like conformation, the α-phosphate is positioned within the H-bond distance (2.9 Å) of Nϵ of Arg206. The conversion between the OtsA- and PimA-like conformers occurs through a rotation of approximately ∼120° about the C4′–C5′ bond of the ribose, drastically altering the position of the α-phosphate and shifting the β-phosphate by about 0.5–0.6 Å away from Arg206, whereas the guanosine moiety remains stationary. Interestingly, the triclinic crystal form shows density only for the PimA-like conformer (Fig. 4A).
Modeling the Ternary Complex
In order to understand the structural basis of regio-selectivity of mannosyl transfer, we modeled the closed configuration of PimB′ because this is assumed to represent the catalytically competent conformation. Attempts to crystallize ternary complexes of PimB′ bound to GDP and acceptor substrate analogs were not successful. Crystals formed readily when (C-terminally tagged) PimB′ was incubated with excess amounts of GDP and of either d-myo-inositol-1-phosphate or deacylated PIM1. Nonetheless, the diffraction data consistently revealed density for the nucleotide but none for the acceptor. Binding studies for PimA (27) and kinetic studies of MshA (46) showed that the substrates bind sequentially, with the sugar-nucleotide binding first, followed by the acceptor. This scenario very likely applies to PimB′ as well, and acceptor binding may require the “closed” configuration, which, in the case of PimB′, appears to be disfavored in crystal lattice formation.
The model of the closed form of the enzyme was constructed using the template of the ternary complex of C. glutamicum MshA (Protein Data Bank entry 3c4v), the closest structural neighbor of PimB′ according to our DALI search. The structures of PimB′ (orthorhombic form) and MshA were aligned by secondary structure matching with respect to their C-terminal domains, followed by aligning the N-terminal domain of PimB′ to the N-terminal domain of MshA, which involves a 27° rotation (Fig. 5A). The rigid body rotary movement of the N-terminal domain toward the C-terminal domain leads to minor steric clashes between helix α5′ and the β11-α11 loop. However, as the comparison between the “triclinic” and “orthorhombic” structures of PimB′ has demonstrated, the α5′ region is conformationally flexible. Therefore, it is justifiable to assume that this region will adapt to spatial constraints imposed by the rotary movement.
Because density for the mannose moiety was not sufficiently defined, we modeled its orientation according to the nucleotide-on-nucleotide superposition of PimB′ with the UDP-glucose bound structure of OtsA. The latter complex structure in conjunction with the ternary complex of OtsA bound to UDP and a disaccharide mimetic inhibitor has provided the most compelling structural evidence as yet to support the proposed SNi internal return mechanism for retaining glycosyltransferases of the GT-B fold (see “Discussion”) (24, 43–45).
Despite restricting the protein modeling to a rigid body motion of the N-terminal domain and positioning the mannose moiety of GDP-Man according to a structural superposition between enzymes that share only 15% sequence identity, the resulting structural model displays a number of mechanistically plausible features. First, the rotary motion places residue Ile18, which precedes the Gly-Gly motif, on the face of the guanosine base, analogous to the interaction of the corresponding residues in PimA (Pro), and OtsA (Ala) (Fig. 5B). The resulting closed binding cavity for the sugar-nucleotide can accommodate GDP-Man without steric overlap. The binding cavity also allows the nucleotide to adopt both conformations that we observed for the pyrophosphate. Importantly, the binding cavity shows a prominent opening, which selectively exposes the C2 hydroxyl and the C1 carbon of GDP-mannose to the putative acceptor-binding site (Fig. 5B).
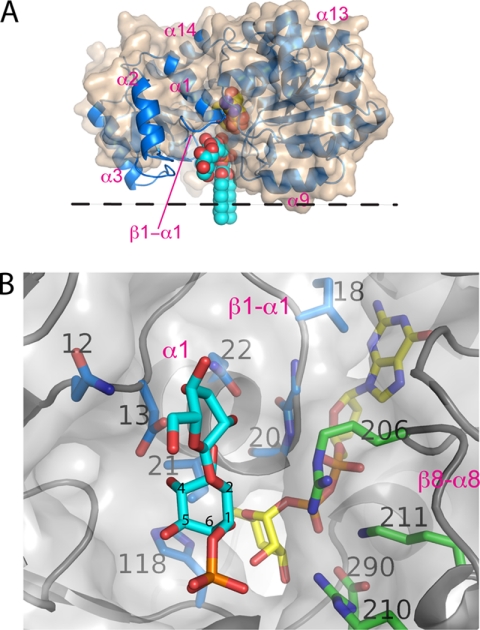
We manually docked a model of PIM1 (2-mannosyl-phosphatidyl-myo-inositol-1-phosphate) into the cleft of the “closed” enzyme (shown in cyan in Fig. 5), such that the 6-hydroxyl is pointing toward the exposed mannose moiety, whereas the mannosylated C2 is pointing in the opposite direction. In this way, the 3- and 4-OH groups of the inositol ring are positioned within hydrogen bonding distance to the carboxylate of Asp13 in the β1-α1 loop, providing a bidentate anchoring interaction that defines the orientation of the inositol ring. In PimA, the residue corresponding to Asp13 is Tyr9, which has been previously implicated in acceptor substrate specificity of mannosyl transfer (10, 14, 27). The phosphate moiety is positioned within 3.5 Å of the guanido group of Arg210, suggesting that Arg210 might contribute to acceptor recognition. We also note that in this model, the position of the phosphate moiety falls in close range of an ordered sulfate molecule of the triclinic structure, which is coordinated by Arg206 (Fig. 3B). The model of the closed complex then served as a guide to probe the structural determinants of acceptor binding by site-directed mutagenesis.
Mutagenesis
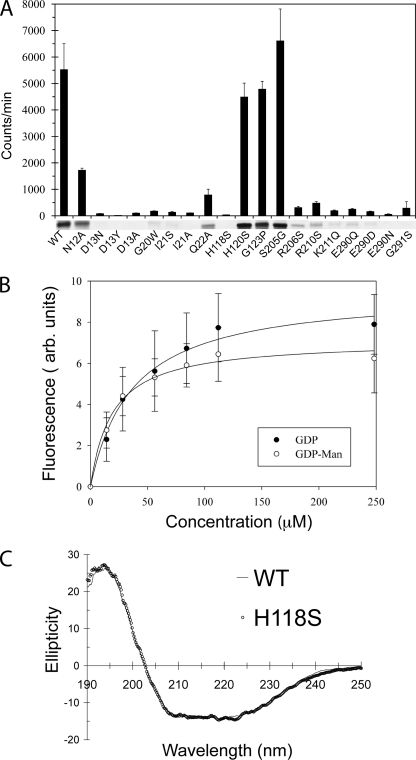
In order to assess the significance of selected residues for PimB′ function, we probed enzyme activity by monitoring the transfer of 14C-labeled mannose from GDP-Man to Ac1PIM1 (Fig. 6A and Table 2). Products were analyzed in terms of radioactivity and by thin layer chromatography (Fig. 6A), whereas nucleotide binding was assayed by intrinsic tryptophan fluorescence (Fig. 6B). In addition, proper folding of mutants was ascertained by circular dichroism spectroscopy (Fig. 6C and supplemental Fig. 4S).
FIGURE 6.
Activity and nucleotide binding of C. glutamicum PimB′ and of selected point mutants. A, analysis of transfer of 14C-labeled mannose from GDP-[14C]Man to acylated phosphatidylinositol mannoside by scintillation counting and thin layer chromatography. B, saturation binding of GDP-Man or GDP to PimB′ monitored by internal Trp fluorescence. C, far UV CD spectra of wild-type PimB′ and of the H118S mutant.
Residues of the C-terminal domain that contact the nucleotide very sensitively influence enzymatic activity. Residual activity of single-residue mutants of the three basic side chains (Arg206, Arg210, and Lys211) situated in the vicinity of the pyrophosphate is less than 9% that of wild-type (Fig. 3B and Table 2). Likewise, mutation of the conserved Glu residue at position 290 to either Gln or Asp reduced activity by more than 95% (Fig. 6A and Table 2). Although detrimental for activity, all of these mutants still bind the acceptor substrate with appreciable affinity. Thus, these mutations generally weaken but do not abrogate binding of GDP or GDP-Man.
Although the nucleotide binding site is highly conserved between PimA and PimB′, subtle structural differences are evident. For instance, Gly291 in PimB′ corresponds to a serine in PimA. Modeling the GDP-Man in PimB′ according to the PimA·GDP-Man complex indicated potential steric hindrance between a serine on position 291 and the mannose moiety. Indeed, compared with wild-type PimB′, the G291S mutant shows 95% reduced activity and a nearly 4-fold increase in Kd for binding of GDP-Man (Table 2). In contrast, the substitution of Ser205 with the corresponding Gly in PimA increases activity by about 10%, although the increase is within one S.D. value.
DISCUSSION
PIM biosynthesis occurs through a series of enzyme-catalyzed reactions during which PI is decorated on the inositol moiety with mannosyl groups in an ordered and regio-selective manner. The present structure in combination with the mutational analysis sheds light on how regio-selectivity is achieved and provides indirect evidence for the hypothesis that catalysis occurs through a substrate-mediated SNi internal return mechanism.
In terms of fold and overall structure, PimB′ conforms to the paradigm of GT-B glycosyltransferases. However, a couple of observations are noteworthy. First, nucleotide binding did not result in crystallization of a closed configuration of the enzyme, in contrast to the closed states observed for the nucleotide-bound complexes of PimA, MshA, OtsA, or WaaG (26, 27, 29, 45). However, it seems improbable that PimB′ is unable to close the active site cleft, given the close similarity to the structural homologs. Small changes in orientation of the domains between the orthorhombic and triclinic crystal form provide direct evidence for conformational flexibility. More importantly, the G20W mutation leads to a 97% drop in activity but binds the donor with an affinity similar to that of wild type PimB′ (Table 2). The fact that a bulky side chain in the center of the cleft is tolerated in terms of nucleotide binding but abrogates activity is consistent with the notion that it impedes the closing motion.
Second, the conformational flexibility observed in the β5-α5 loop of the active site cleft has not been observed previously in GT4 glycosyltransferases. The shift of individual side chains and the corresponding change of intramolecular contacts seem significant. For instance, the position of the Trp124 Cα is shifted by 7 Å, and its side chain makes contacts with different parts of the structure in the two crystal forms. We probed whether helix α5′ is required for activity by substituting glycine at position 123 with proline, but the resulting drop of activity is only about 15% (Table 2). Furthermore, His120 at the N terminus of helix α5′ is only ordered in the triclinic structure and disordered when helix α5′ forms. Substituting His120 by serine reduced activity by about 20% (Table 2), whereas the CD spectrum of the mutant G123P is virtually identical to that of wild-type PimB′ (supplemental Fig. 4S). In light of these data, we hypothesize that the conformational flexibility of the β5-α5 loop works in concert with the opening/closing motion of the active site cleft upon substrate binding but influences catalysis only in an indirect fashion if at all.
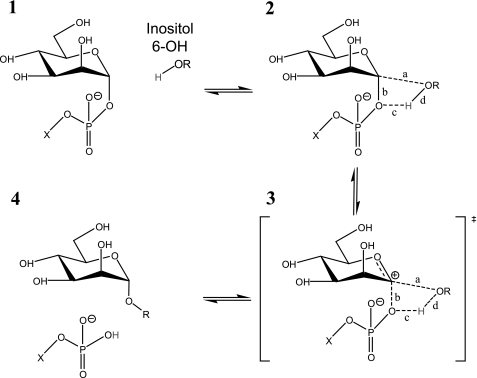
PimB′ is a GT-B glycosyltransferase that retains the stereochemical configuration of the anomeric carbon of the donor saccharide. The catalytic reaction mechanism of retaining GT-B transferases has remained something of a mystery. In glycoside hydrolases, retention of the anomeric configuration occurs through an SN2-like double displacement mechanism, mediated by a pair of side chain carboxylates and involving a covalent sugar-enzyme intermediate (47). However, none of the inhibitor- or substrate mimic-bound complexes solved to date has provided structural evidence to support such a scenario in GT-B glycosyltransferases (24, 48–50). Instead, a substrate-mediated SNi (internal return) mechanism has been invoked (reviewed in Ref. 24). This mechanism involves three tightly linked reaction steps (Scheme 1): first, decay of the sugar-phosphate bond, leaving a (solvent-separated) ion pair of the oxocarbenium ion-like saccharide and the negatively charged phosphate (panels 2 and 3); second, deprotonation of the acceptor hydroxyl by the phosphate leaving group; third, nucleophilic attack by the acceptor on the oxocarbenium ion (panels 3 and 4). The first step is rate-limiting, whereas the subsequent steps must occur on a time scale shorter than solvent attack or ion pair reorganization (24). If correct, this mechanism imposes constraints on the geometry of donor and acceptor substrate in the ternary complex, in that the leaving group must be sufficiently close to the acceptor to be able to deprotonate the attacking acceptor hydroxyl.
SCHEME 1.
Schematic representation of steps of the SNi-like (internal return) reaction mechanism leading to retention of the α-configuration of the anomeric carbon of the donor saccharide. This scheme was adapted from Ref. 43.
In our model of the acceptor-bound complex, the 6-OH of inositol (Oi6) is positioned within ∼4 Å of the C1 carbon of the donor mannose and within 5 Å of the β-phosphate. The distance vectors Oi6-C1 and C1-β-phosphate enclose an angle of ∼105°. Thus, the geometric configurations of the substrates in the ternary complex model are compatible with the requirements of the SNi reaction mechanism. Our mutagenesis data provide further supporting evidence for a substrate-dependent reaction mechanism. For instance, the most obvious candidate to act as a catalytic nucleophile in an SN2-like mechanism is Glu290, a carboxylate that is strictly conserved across GT4 family enzymes and positioned close (∼4.5 Å) to the sugar-phosphate bond. Switching this residue to glutamine, which should completely impair nucleophilic attack, decreases activity by about 95% while leaving substrate binding largely intact (<3-fold increase of Kd). Importantly, the residual activity is about 10 times above base-line level and can be reduced further by the E290D substitution (Table 2). In contrast, the substitution of highly conserved His118 by serine decreases activity essentially to base-line level. In the “open” configuration, His118 does not contact the donor substrate, but in the “closed” model, it contacts the C6 hydroxyl of the nucleotide-linked mannose. His118 is positioned closely opposite to Ile21 (closest contact 3.9 Å). Substitution of Ile21 with either Ser or Ala diminishes activity to about 2.5%. The close proximity of a hydrophobic side chain to His118 suggests that the latter is pushed to its non-ionized state, which could enable the His side chain to act as a nucleophile. However, in our modeled ternary complex as in the experimental structures of OtsA (Protein Data Bank entry 2wtx) or MshA (Protein Data Bank entry 3c4v), the position of His118 relative to the nucleotide makes such a role improbable.
In their structural evaluation of PimA, Guerin et al. (27) identified Tyr9 as a residue that might form a stacking interaction with the inositol ring of PI and thus may be key to acceptor specificity. This tyrosine is located in the β1-α1 loop and corresponds to Asp13 in PimB′. Within the sequences of PimB′ and PimA enzymes, respectively, the Asp and Tyr are strictly conserved at this position (supplemental Fig. 1S). In the model of the ternary complex, Asp13 is positioned within H-bond distance to the 3- and 4-OH of the inositol moiety (i.e. the only inositol hydroxyls that are in an equatorial configuration). Such a bidentate interaction would ensure that inositol presents the axial 6-OH rather than the 2-OH to the C1 carbon of mannose. Indeed, swapping the positions of 3-OH and 4-OH of inositol would present the 5-OH to C1 of mannose rather than 2-OH. Probing Asp13 by mutagenesis, it is intriguing that even the most conservative substitution, aspartate to asparagine, reduces activity by 98.5% relative to wild type. Indeed, this subtle change knocks down activity as effectively as the more drastic Asp to Ala substitution (Table 2). Residues in the immediate vicinity (Asn12, Ile21, and Gln22) are similarly sensitive to substitutions, causing a reduction of activity by at least 70%. We also probed whether the specificity of PimB′ can be switched to mannosylation of the 2-OH by substituting Asp13 by tyrosine. However, the result was a knockdown of activity to blank level (Table 2). Thus, residues in the β1-α1 loop play a key role in acceptor recognition, but the single substitution on position 13 is not sufficient to switch acceptor specificity.
The deleterious effect of the D13N mutation stresses the significance of Asp13 in determining acceptor specificity, although the mechanistic implication of this substitution is not clear because an asparagine should still be able to form H-bonds with the inositol hydroxyl groups. However, the effect is reminiscent of an analogous mutation in glycosidase family GH68. There, substitution of a strictly conserved Asp by Asn, which formed strong H-bonds with saccharide substrate hydroxyls but, for geometric reasons, could not be a nucleophile or general acid/base, reduced activity by 2 orders of magnitude in terms of kcat. This effect was attributed to stabilization of the oxocarbenium ion transition state by the negatively charged carboxylate (51, 52). Lack of sequence conservation across GT4 family enzymes appears to rule out a fundamental role of Asp13 in a general reaction mechanism of GT4 enzymes. However, we note that PimA extends a glutamic acid side chain (residue 199) toward the inositol ring when superimposed with our model of the PimB′ ternary complex. This glutamic acid, which is critical for activity (10), is located in the β8-α8 loop and, in the structural superposition, lines up with Val208 of PimB′. Our model suggests that Glu199 could form H-bond contacts with the equatorial OH groups of inositol if the inositol moiety is oriented such that the 2-OH is presented to the C1 of the donor saccharide. Thus, the active site cleft of PimA includes a side chain that could play a role functionally analogous to that of Asp13 in PimB′.
The dual conformation of the pyrophosphate in complexes co-crystallized with GDP-Man is intriguing. This feature could be mechanistically linked to release of the donor mannosyl group following decay of the sugar-phosphate bond and transfer to inositol. PimB′·GDP complexes have a pronounced preference for the “OtsA-like” nucleotide conformation, whereas GDP-Man bound PimB′ displayed either the superposition of the OtsA- and PimA-like states or only the PimA-like state. Density for the mannose group is completely missing in the orthorhombic complex. However, the presence of a disordered sugar moiety can be inferred on the one hand from the pronounced preference for a dual nucleotide conformation in the GDP-Man co-crystals and, second, from the positioning of an extra water molecule between β-phosphate and Glu290 in the GDP co-crystals (W4 in Fig. 4C). These observations were consistent between at least two different crystals of each complex. Between the PimA- and OtsA-like conformations, the β-phosphate shifts away from Arg206 by about 0.5–0.6 Å, weakening the corresponding ionic interaction, which would facilitate increased mobility of Arg206. Such increased mobility of this side chain could promote release of the transferred mannosyl group from the confines of the donor substrate-binding cavity.
Conclusions
Biosynthesis of PIM occurs through a series of glycosyl transfer reactions that initially take place at the cytoplasmic side of the membrane. The final product is presented on the extracellular face of the membrane, and hence flippase-mediated membrane translocation of either the final or an intermediate product must occur along the way (3). We consider the following scenario probable. PimA, PimB′, and PimC act on acceptor substrates (PI and intermediate forms of PIM) that are anchored in the cytoplasmic leaflet of the membrane. The active site geometry of PimB′ and its electrostatic surface potential (supplemental Fig. 5S) are compatible with the notion of transient membrane association, potentially in the orientation shown in Fig. 5A. Liposome binding data recorded for PimA further support this scenario (27). PimB′ differs from PimA in several structural details that reflect their distinct acceptor substrate specificities. Chief among them is the lack of the extended β3-α3 loop, which contributes to acceptor binding in PimA but, based on geometry, cannot play such a role in PimB′. Second, the active site cleft shows a more open surface, to accommodate the mannose attached to the 2-position of inositol. Third, we show that Asp13, which is critical for activity, cannot be swapped for the corresponding tyrosine in PimA. We postulate that Asp13 is chiefly responsible for regio-specific orientation of inositol relative to the donor saccharide. Our mutagenesis data also lend indirect support to the hypothesis that glycosyl transfer with retention of the α-configuration of the anomeric carbon occurs through a substrate-mediated SNi-type mechanism.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1S–5S.
The atomic coordinates and structure factors (codes 3oka, 3okc, and 3okp) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PIM
- phosphatidylinositol mannoside
- PI
- phosphatidyl-myo-inositol-1-phosphate
- GDP-Man
- guanosine-5′-diphosphate-α-d-mannose
- LM
- lipomannan
- LAM
- lipoarabinomannan
- GT4
- glycosyltransferase family 4
- NCS
- non-crystallographic symmetry
- r.m.s.
- root mean square
- SeMet
- selenomethionine
- H-bond
- hydrogen bond
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Józefowski S., Sobota A., Kwiatkowska K. (2008) BioEssays 30, 943–954 [DOI] [PubMed] [Google Scholar]
- 2.Guenin-Macé L., Siméone R., Demangel C. (2009) Transbound Emerg. Dis. 56, 255–268 [DOI] [PubMed] [Google Scholar]
- 3.Berg S., Kaur D., Jackson M., Brennan P. J. (2007) Glycobiology 17, 35–56R [DOI] [PubMed] [Google Scholar]
- 4.Doz E., Rose S., Court N., Front S., Vasseur V., Charron S., Gilleron M., Puzo G., Fremaux I., Delneste Y., Erard F., Ryffel B., Martin O. R., Quesniaux V. F. (2009) J. Biol. Chem. 284, 23187–23196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhoades E. R., Archambault A. S., Greendyke R., Hsu F. F., Streeter C., Byrd T. F. (2009) J. Immunol. 183, 1997–2007 [DOI] [PubMed] [Google Scholar]
- 6.Fischer K., Scotet E., Niemeyer M., Koebernick H., Zerrahn J., Maillet S., Hurwitz R., Kursar M., Bonneville M., Kaufmann S. H., Schaible U. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10685–10690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Salle H., Mariotti S., Angenieux C., Gilleron M., Garcia-Alles L. F., Malm D., Berg T., Paoletti S., Maître B., Mourey L., Salamero J., Cazenave J. P., Hanau D., Mori L., Puzo G., De Libero G. (2005) Science 310, 1321–1324 [DOI] [PubMed] [Google Scholar]
- 8.Driessen N. N., Ummels R., Maaskant J. J., Gurcha S. S., Besra G. S., Ainge G. D., Larsen D. S., Painter G. F., Vandenbroucke-Grauls C. M., Geurtsen J., Appelmelk B. J. (2009) Infect. Immun. 77, 4538–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahon R. N., Rojas R. E., Fulton S. A., Franko J. L., Harding C. V., Boom W. H. (2009) Infect. Immun. 77, 4574–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerin M. E., Schaeffer F., Chaffotte A., Gest P., Giganti D., Korduláková J., van der Woerd M., Jackson M., Alzari P. M. (2009) J. Biol. Chem. 284, 21613–21625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korduláková J., Gilleron M., Mikusova K., Puzo G., Brennan P. J., Gicquel B., Jackson M. (2002) J. Biol. Chem. 277, 31335–31344 [DOI] [PubMed] [Google Scholar]
- 12.Korduláková J., Gilleron M., Puzo G., Brennan P. J., Gicquel B., Mikusová K., Jackson M. (2003) J. Biol. Chem. 278, 36285–36295 [DOI] [PubMed] [Google Scholar]
- 13.Lea-Smith D. J., Martin K. L., Pyke J. S., Tull D., McConville M. J., Coppel R. L., Crellin P. K. (2008) J. Biol. Chem. 283, 6773–6782 [DOI] [PubMed] [Google Scholar]
- 14.Guerin M. E., Kaur D., Somashekar B. S., Gibbs S., Gest P., Chatterjee D., Brennan P. J., Jackson M. (2009) J. Biol. Chem. 284, 25687–25696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra A. K., Batt S., Krumbach K., Eggeling L., Besra G. S. (2009) J. Bacteriol. 191, 4465–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaeffer M. L., Khoo K. H., Besra G. S., Chatterjee D., Brennan P. J., Belisle J. T., Inamine J. M. (1999) J. Biol. Chem. 274, 31625–31631 [DOI] [PubMed] [Google Scholar]
- 17.Tatituri R. V., Illarionov P. A., Dover L. G., Nigou J., Gilleron M., Hitchen P., Krumbach K., Morris H. R., Spencer N., Dell A., Eggeling L., Besra G. S. (2007) J. Biol. Chem. 282, 4561–4572 [DOI] [PubMed] [Google Scholar]
- 18.Mishra A. K., Klein C., Gurcha S. S., Alderwick L. J., Babu P., Hitchen P. G., Morris H. R., Dell A., Besra G. S., Eggeling L. (2008) Antonie Leeuwenhoek 94, 277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kremer L., Gurcha S. S., Bifani P., Hitchen P. G., Baulard A., Morris H. R., Dell A., Brennan P. J., Besra G. S. (2002) Biochem. J. 363, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita Y. S., Sena C. B., Waller R. F., Kurokawa K., Sernee M. F., Nakatani F., Haites R. E., Billman-Jacobe H., McConville M. J., Maeda Y., Kinoshita T. (2006) J. Biol. Chem. 281, 25143–25155 [DOI] [PubMed] [Google Scholar]
- 21.Morita Y. S., Patterson J. H., Billman-Jacobe H., McConville M. J. (2004) Biochem. J. 378, 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson J. H., Waller R. F., Jeevarajah D., Billman-Jacobe H., McConville M. J. (2003) Biochem. J. 372, 77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., Mushegian A. (2003) Protein Sci. 12, 1418–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 25.Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003) J. Mol. Biol. 328, 307–317 [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Fleites C., Proctor M., Roberts S., Bolam D. N., Gilbert H. J., Davies G. J. (2006) Chem. Biol. 13, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 27.Guerin M. E., Kordulakova J., Schaeffer F., Svetlikova Z., Buschiazzo A., Giganti D., Gicquel B., Mikusova K., Jackson M., Alzari P. M. (2007) J. Biol. Chem. 282, 20705–20714 [DOI] [PubMed] [Google Scholar]
- 28.Ruane K. M., Davies G. J., Martinez-Fleites C. (2008) Proteins 73, 784–787 [DOI] [PubMed] [Google Scholar]
- 29.Vetting M. W., Frantom P. A., Blanchard J. S. (2008) J. Biol. Chem. 283, 15834–15844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chua T. K., Bujnicki J. M., Tan T. C., Huynh F., Patel B. K., Sivaraman J. (2008) Plant Cell 20, 1059–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 33.Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 34.CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 35.de la Fortelle E., Bricogne G. (1997) Methods Enzymol. 276, 472–494 [DOI] [PubMed] [Google Scholar]
- 36.Collier A., Wagner G. K. (2008) Chem. Commun. 178–180 [DOI] [PubMed] [Google Scholar]
- 37.Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2007) Curr. Protoc. Protein Sci. Chapter 2, Unit 2.9 [DOI] [PubMed] [Google Scholar]
- 38.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 39.Dobson G., Minnikin D. E., Minnikin S. M., Parlett J. H., Goodfellow M., Magnusson M. (1985) in Chemical Methods in Bacterial Systematics (Goodfelow M., Minnikin D. E. eds) pp. 237–265, Academic Press, London [Google Scholar]
- 40.Besra G. S., Morehouse C. B., Rittner C. M., Waechter C. J., Brennan P. J. (1997) J. Biol. Chem. 272, 18460–18466 [DOI] [PubMed] [Google Scholar]
- 41.Rao S. T., Rossmann M. G. (1973) J. Mol. Biol. 76, 241–256 [DOI] [PubMed] [Google Scholar]
- 42.Holm L., Sander C. (1996) Science 273, 595–603 [DOI] [PubMed] [Google Scholar]
- 43.Errey J. C., Lee S. S., Gibson R. P., Martinez Fleites C., Barry C. S., Jung P. M., O'Sullivan A. C., Davis B. G., Davies G. J. (2010) Angew Chem. Int. Ed. Engl. 49, 1234–1237 [DOI] [PubMed] [Google Scholar]
- 44.Gibson R. P., Tarling C. A., Roberts S., Withers S. G., Davies G. J. (2004) J. Biol. Chem. 279, 1950–1955 [DOI] [PubMed] [Google Scholar]
- 45.Gibson R. P., Turkenburg J. P., Charnock S. J., Lloyd R., Davies G. J. (2002) Chem. Biol. 9, 1337–1346 [DOI] [PubMed] [Google Scholar]
- 46.Newton G. L., Ta P., Bzymek K. P., Fahey R. C. (2006) J. Biol. Chem. 281, 33910–33920 [DOI] [PubMed] [Google Scholar]
- 47.Zechel D. L., Withers S. G. (2001) Curr. Opin. Chem. Biol. 5, 643–649 [DOI] [PubMed] [Google Scholar]
- 48.Sheng F., Jia X., Yep A., Preiss J., Geiger J. H. (2009) J. Biol. Chem. 284, 17796–17807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steiner K., Hagelueken G., Messner P., Schäffer C., Naismith J. H. (2010) J. Mol. Biol. 397, 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frantom P. A., Coward J. K., Blanchard J. S. (2010) J. Am. Chem. Soc. 132, 6626–6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng G., Fütterer K. (2003) Nat. Struct. Biol. 10, 935–941 [DOI] [PubMed] [Google Scholar]
- 52.Yanase H., Maeda M., Hagiwara E., Yagi H., Taniguchi K., Okamoto K. (2002) J. Biochem. 132, 565–572 [DOI] [PubMed] [Google Scholar]
- 53.Engh R., Huber R. (1991) Acta Crystallogr. A 47, 392–400 [Google Scholar]
- 54.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]