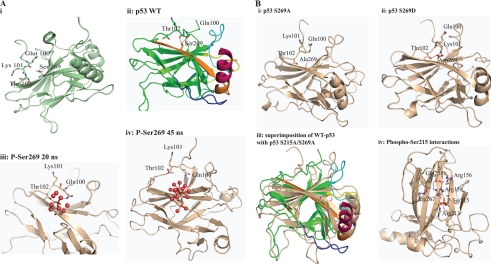
FIGURE 11.
Model summarizing the effect of phosphorylation at the conformationally flexible motif on p53 activity. A, i, diagram of the structure of the wild type p53 core domain (Protein Data Bank entry 2FEJ) highlighting the Ser269 region and its proximity to the flexible motif (residues 100–103). ii, the epitope is in orange color, whereas the loop and helix that interact with the DNA are shown in yellow and brown, respectively. ii–iv, molecular dynamics modeling snapshots of p53 mutants to give rise to mechanisms of p53 unfolding over time. Shown are phospho-Ser269 effects on p53. ii, wild type p53 highlighting interactions of Ser269 with Thr102 and Gln100; iii, the phospho-Ser269 intermediate; iv, phospho-Ser269. Because no stabilizing cationic side chains exist near the phosphate, the negatively charged phosphate is energetically more stable when solvated by water. This is accompanied by the movement away of the N-terminal loop and destabilization of the DNA binding regions plus the dimerization interface. B, molecular dynamics modeling snapshots of p53 mutants to give rise to mechanisms of p53 unfolding over time. Shown are Asp269 or Ala269 effects on p53. i, S269A final molecular dynamics snapshot; ii, S269D final molecular dynamics snapshot; iii, superposition of wild type and double alanine final molecular dynamics snapshot; iv, molecular dynamics modeling snapshots of p53 mutants to give rise to mechanisms of p53 unfolding over time. Shown are phospho-Ser215 effects on p53 and the phospho-Ser215 intermediate showing the network of interactions. Phosphorylation at Ser215 leads to the formation of an extended network that involves Arg213-Ser(P)215-Arg158-Asp258-Arg156.