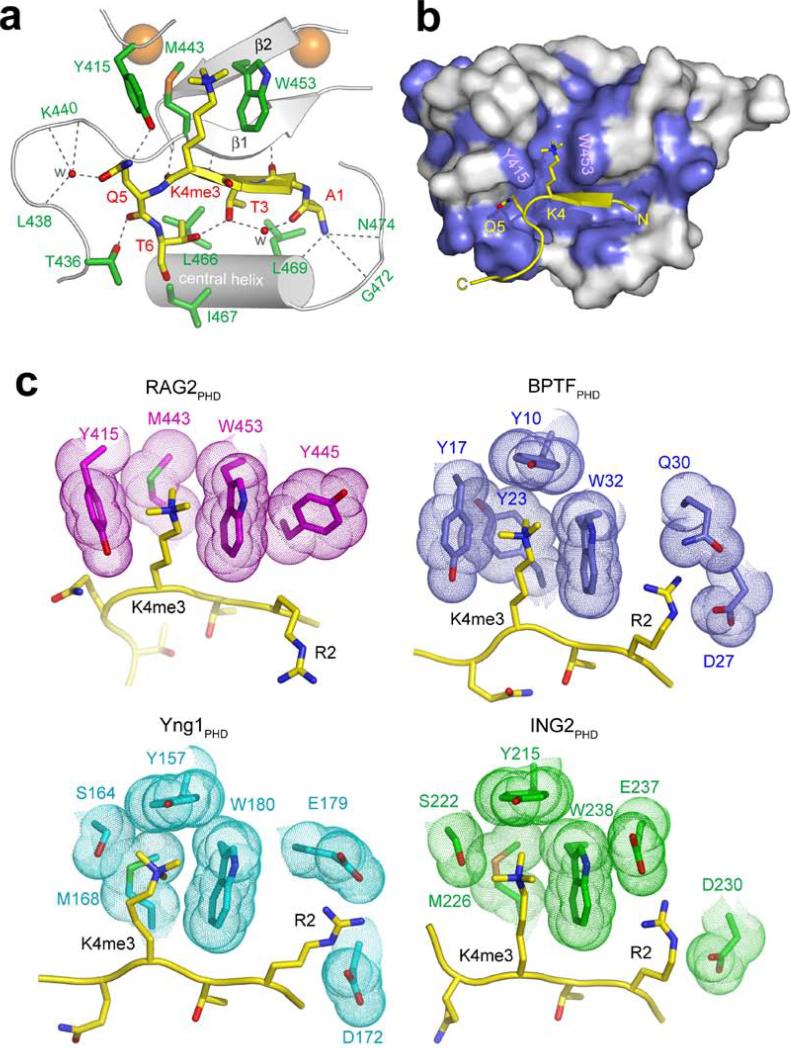
Fig. 2. Molecular Basis of H3K4me3-recognition by RAG2PHD.
a, b, 1.15 Å crystal structure of RAG2414-487 complexed with H3K4me3 peptide. a, Ribbon diagram of the complex. For clarity, only the central portion of RAG2414-487 is shown (silver). Residues whose side chains interact with the H3 peptide are shown in green sticks with blue (nitrogen) and red (oxygen) highlights. Residues whose main chain atoms interact with the peptide are labeled in green. The peptide is shown in yellow. With the exception of R2, the side chains of A1 to T6 all interact with RAG2 and are shown in stick models. Zn2+ ions are shown as orange spheres and water molecules mediating protein-peptide interactions are shown as red spheres. Grey dashed lines represent hydrogen bonds. RAG2PHD consists of two unorthodox, interdigitated zinc fingers, linked by a pair of anti-parallel β-strands and a central α-helix. The backbone of the first four residues of the histone H3 peptide are hydrogen bonded with one of the β-strands of RAG2PHD, forming a 3-stranded antiparallel β-sheet. b, The H3 peptide binding surface is conserved among RAG2 proteins. Residues of RAG2 that are conserved through evolution (see Fig. S5) are colored blue on the molecular surface. c, Structural comparison of the PHD fingers from RAG2, BPTF, Yng1 and ING2. Side chains in the PHD fingers that interact with H3K4me3 and H3R2 are highlighted with molecular surface.