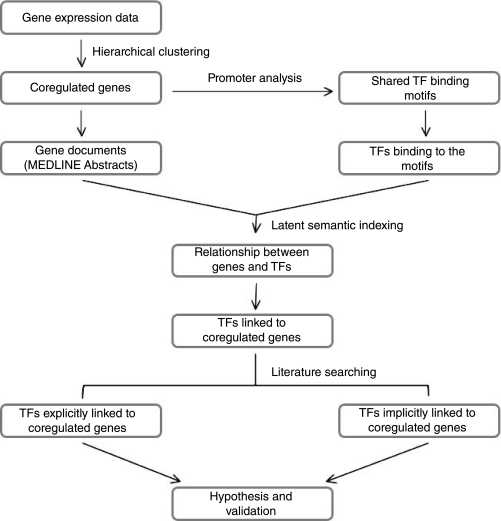
FIG. 1.
Overview of the method used to identify candidate regulatory transcription factors (TFs) from microarray data. Small groups of genes that are similarly affected by experimental treatments are selected for promoter sequence analysis to identify shared TF-binding motifs. A matrix consisting of pairwise literature relationships between each TF and co-regulated gene is constructed by Latent Semantic Indexing (LSI). The relationship dvalues for genes and TFs are then subjected to hierarchical clustering and the TFs that have high literature relationship to the co-regulated genes are selected for manual analysis of the literature to determine an explicit or implicit relationship.