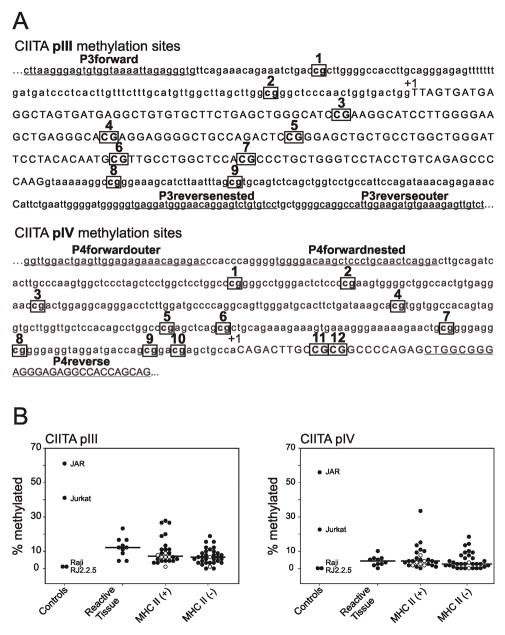
Figure 1.
(A) CIITA promoter sites tested for CpG methylation and primers used for nested PCR. Possible CpG sites for methylation of CIITA promoters pIII and pIV are boxed and numbered. Sites for primer binding are underlined. Exons are in capitals and indicated by +1 [8]. (B) Percentage methylation of CIITA promoters pIII and pIV. Tallies of methylation marks within each of the ten amplicons per sample, per promoter, were divided by the number of possible methylation marks in ten samples (i.e., 90 for pIII, and 120 for pIV), to give a percentage methylation for each sample. Raji and RJ2.2.5 do not demonstrate CIITA methylation, as described in the text, and were used as negative controls. Jurkat and JAR demonstrate CIITA methylation, as described in the text, and were used as positive controls. Kruskal-Wallis One Way Analysis of Variance on Ranks and Dunn’s method were used to determine significant differences in methylation of CIITA pIII and pIV between reactive tissues, MHCII(+), and MHCII(−) samples using SigmaPlot 11.0 (San Jose, CA). Cell lines within the MHCII(+) and MHCII(−) groups are indicated with open circles.